Abstract
The complete mitochondrial (mt) genome was sequenced in two specimens of the plain sculpin Myoxocephalus jaok by high-throughput sequencing technology (Ion S5 platform). The sequences were 16,653 bp in size, and the gene content, gene arrangement, and size were very similar to other sculpin mt genomes published previously. Comparison of the two M. jaok mt genomes now obtained with other complete mt genomes available in GenBank reveals an affinity to the sculpin fishes from the genus Myoxocephalus. The low level of sequence divergence (1.6%) detected between the genome of M. jaok and the GenBank complete mitochondrial genomes of Myoxocephalus polyacanthocephalus (MK621914, MK621915) may likely be due to recent divergence of the species and/or historical hybridization.
The plain sculpin Myoxocephalus jaok (Cuvier) has a wide distribution in the North Pacific including the Sea of Japan, the Sea of Okhotsk, the Bering Sea, the Chukchi Sea, eastern Aleutian Islands, and Gulf of Alaska (e.g. Neyelov Citation1979; Lindberg and Krasyukova Citation1987; Fedorov et al. Citation2003; Mecklenburg et al. Citation2016). The species is characterized by variable morphology, which makes taxonomic identification difficult. Genetic data for M. jaok are scarce (Podlesnykh and Moreva Citation2014; Moreva et al. Citation2017); the complete mitochondrial (mt) genome of M. jaok had not yet been sequenced. To increase the power of phylogenetic analysis of this complex fish group we have sequenced two complete mt genomes of M. jaok (GenBank accession numbers MN162694 and MN162695) from the Sea of Japan, Sakhalin Island (sample MJA4-17; Aug-07-2017; 50.89277°N 142.10077°E) and the Sea of Japan, Chikhachyova Bay (sample MJA5-17; Aug-09-2017; 51.48575°N 140.8244°E). Fish specimens are stored at the museum of the A. V. Zhirmunsky National Scientific Center of Marine Biology, Vladivostok, Russia (www.museumimb.ru) under accession numbers MIMB 37998 and MIMB 37999.
The genomic DNA was extracted using the KingFisher Flex System and a set of reagents MagMAX DNA Multi-Sample Kit (ThermoFisher Scientific, Waltham, MA, USA). The complete mt genomes were amplified in five overlapping fragments using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific). Libraries were prepared using Ion Plus Fragment Library Kit and unique adapters (Ion Xpress,Waltham, MA, USA) with pre-fragmentation on the focused ultrasonicator Covaris M220. Ready libraries were sequenced on the Ion S5 sequencing platform (ThermoFisher Scientific) at the Far Eastern Federal University (Vladivostok, Russia). The complete mt genomes obtained were initially annotated using the MitoFish Web Server (Iwasaki et al. Citation2013) and further manually adjusted with MEGA 7 (Kumar et al. Citation2016) by comparisons with mt genomes of other sculpin fishes.
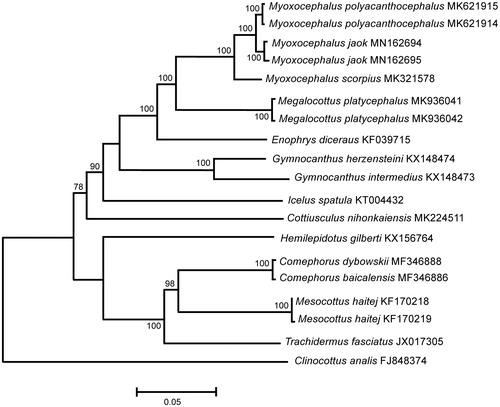
The M. jaok mt genomes (120× coverage) are 16,653 bp in size; the gene arrangement, composition, and size are very similar to the sculpin fish genomes published previously. There are 44 single nucleotide differences between the two genomes; total sequence divergence (Dxy) is 0.0026 ± 0.0004. Comparison of the two mt genomes now obtained with other complete mt genomes available in GenBank for the genera Myoxocephalus, Megalocottus, Enophrys, Icelus, Gymnocanthus, Cottiusculus, Hemilepidotus, Mesocottus, Comephorus, Trachidermus, and Clinocottus reveals an affinity of M. jaok to the sculpin fishes of the genus Myoxocephalus (). The difference (Dxy) between M. jaok and Myoxocephalus polyacanthocephalus is low, 0.0160 ± 0.0010, and could be explained by recent divergence and/or hybridization. Very low divergence (Dxy = 0.0070 ± 0.0030) is also detected between other sculpin species (), Comephorus baicalensis (MF346886) and Comephorus dybowskii (MF346888). Moreover, using the mt sequences (COI, CYTB, and 16S rRNA), Podlesnykh and Moreva (Citation2014) and Moreva et al. (Citation2017) detected low divergence (1.54 − 2.08%) between M. jaok and Myoxocephalus stelleri, which is significantly lower than the average interspecies value, 0.0380 ± 0.020, obtained previously for sculpin fishes based on the COI marker (Kartavtsev et al. Citation2009). Thus, the 5′-COI ‘barcode’ region is not representative to characterize taxonomic relationships between sculpin fishes.
Figure 1. Maximum-likelihood tree for the plain sculpin Myoxocephalus jaok and GenBank representatives of the family Cottidae. The tree is constructed using whole mitochondrial genome sequences. The tree is based on the Hasegawa–Kishino–Yano + gamma + invariant sites (HKY + G+I) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Fedorov VV, Chereshnev IA, Nazarkin MV, Shestakov AV, Volobuev VV. 2003. Catalog of marine and freshwater fishes from the northern sea of Okhotsk. Vladivostok (Russia): Dalnauka.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kartavtsev YPh, Sharina SN, Goto T, Balanov AA, Hanzawa N. 2009. Sequence diversity at cytochrome oxidase 1 (Co-1) gene among sculpins (Scorpaeniformes, Cottidae) and some other scorpionfish of Russia Far East with phylogenetic and taxonomic insights. Genes Genom. 31(2):183–197.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lindberg GU, Krasyukova ZV. 1987. Fishes of the Sea of Japan and the adjacent areas of the sea of Okhotsk and the Yellow Sea. Part 5: Scorpaeniformes. Leningrad (Russia): Nauka.
- Mecklenburg CW, Mecklenburg TA, Sheiko BA, Steinke D. 2016. Pacific Arctic marine fishes. Conservation of Arctic Flora and Fauna Monitoring Series Report no. 23. Akureyri (Iceland): CAFF International Secretariat.
- Moreva IN, Radchenko OA, Petrovskaya AV, Borisenko SA. 2017. Molecular genetic and karyological analysis of antlered sculpins of Enophrys diceraus group (Cottidae). Russ J Genet. 53(9):1030–1041.
- Neyelov AV. 1979. Seismosensory system and classification of sculpins (Cottidae: Myoxocephalinae, Artedellinae). Leningrad (Russia): Nauka.
- Podlesnykh AV, Moreva IN. 2014. Variability and relationships of the Far Eastern species of sculpins Myoxocephalus and Megalocottus (Cottidae) based on mtDNA markers and karyological data. Russ J Genet. 50(9):949–956.
