Abstract
Chrysopogon aciculatus (Retz.) Trin. is one of the most important warm-season turfgrass and one of the distribution centers in China, widely distributed in tropical regions of South China. In this study, the complete chloroplast genome sequence of C. aciculatus was determined from Illumina pair-end sequencing data. The complete chloroplast genome sequence of C. aciculatus is 140,696 base pairs (bp) in length, including one large single-copy region (LSC, 82,667 bp), one small single-copy region (SSC, 12,539 bp), and a pair of inverted repeat regions (IRs) of 22,745 bp. Besides, the complete chloroplast genome contains 134 genes, including 85 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that C. aciculatus (Retz.) Trin. was most closely related to C. serrulatus. The complete chloroplast genome will provide reference for the further investigation and research of C. aciculatus.
The perennial grass genus Chrysopogon is mainly distributed in tropical and subtropical regions of the world. Chrysopogon aciculatus (Retz.) Trin. is the only member of the genus suitable for use as a turfgrass (Zhang et al. Citation2016). Chrysopogon aciculatus is widely distributed in southern China and commonly used in lawns, sports fields, and shaded turf (Zheng et al. Citation2005). Previous studies of C. aciculatus have mainly focused on pollination (Reddi et al. Citation2010), morphological characteristics (Liao et al. Citation2011), taxonmy (Ambasta and Rana Citation2013), molecular genetic markers (Zhang et al. Citation2016), etc. To date, the complete chloroplast genome of C. aciculatus is little known. Therefore, This research will contribute to the species identification, germplasm diversity, and genetic engineering of C. aciculatus.
The plant material of C. aciculatus was collected from Hainan university Danzhou Campus, Haikou, Hainan Province, China. The voucher specimen is kept at Hainan University (specimen code HD20160816). The total genomic DNA was extracted from fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. Approximately 5 Gb of sequenced data were extracted from the total sequencing output and input into Organelle PBA (Soorni et al. Citation2017) to assemble the chloroplast genome. Annotation of the chloroplast genome was performed using the Dual Organellar GenoMe Annotator (DOGMA) online tool (Wyman et al. Citation2004) and Geneious v11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012), then manually verified and corrected by comparison with C. serrulatus (GenBank accession NC_029884.1). Finally, we obtained a complete chloroplast genome of C. aciculatus and submitted to GenBank with an accession number (MN974486).
The complete plastid genome sequence of C. aciculatus is 140,696 bp in length with 82,667 bp of the large single-copy (LSC) region, 12,539 bp of the small single-copy (SSC) region, and 22,745 bp of the invert repeats (IR) regions. The overall GC content of the plastid genome was 38.39%, while the corresponding values of the LSC, SSC, and IR regions were 36.23%, 32.67%, and 43.9%, respectively. The genome contained 134 genes, 85 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Among those, seven protein-coding genes (ndhB, rpl2, rpl23, rps7, rps15, rps19, and ycf68), eight tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), and four rRNA genes were duplicated in IR regions. Fourteen genes (eight protein-coding genes and six tRNA genes) contained one intron, and two genes (rps12 and ycf3) contained two introns.
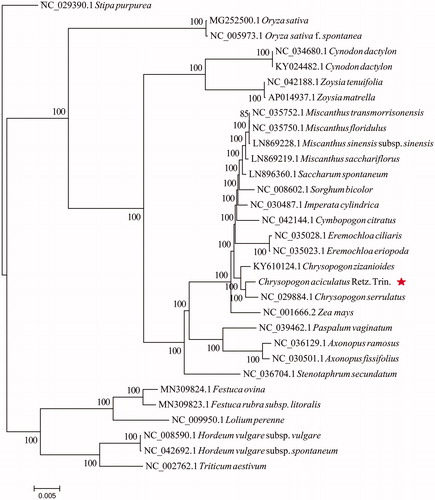
To reveal the phylogenetic position of C. aciculatus with other 30 species of the Poaceae, a molecular phylogenetic tree was constructed based on complete chloroplast genome sequences. All the sequences were downloaded from NCBI GenBank. Alignment was conducted using MAFFT v7.307 (Katoh and Standley Citation2013). The phylogenetic tree was built using RAxML (Stamatakis Citation2014) with bootstrap set to 1000. Stipa purpurea (NC_029390.1) served as the outgroup. The maximum likelihood(ML)phylogenetic tree suggested that C. aciculatus (Retz.) Trin. is closely related to C. serrulatus (). The results will not only help to investigate population genetic diversity of C. aciculatus, but also contribute to the molecular phylogeny studies of Chrysopogon.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ambasta N, Rana NK. 2013. Taxonomical study of Chrysopogon aciculatus (Retz.) Trin. a significant grass of Chauparan, Hazaribag(Jharkhand). Sci Res Rep. 3:27.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liao L, Bai CJ, Guo XL, Wang ZY. 2011. Morphological diversity of Chrysopogon aciculatus. Chinese J Trop Crops. 32:2042–2047.
- Reddi CS, Raju NSN, Rao M. 2010. Pollination and seed set in tropical wetland grasses. Nord J Bot. 28:354–365.
- Soorni A, Haak D, Zaitlin D, Bombarely A. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18(1):49.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang XY, Liao L, Liu Y, Wang ZY, Liu JX. 2016. Isolation and characterization of new polymorphic simple sequence repeat loci in Chrysopogon aciculatus (Retz.) Trin. HortScience. 51(3):232–235.
- Zhang XY, Liao L, Wang ZY, Bai CJ, Liu JX. 2016. Analysis of genetic diversity in Chrysopogon aciculatus using intersimple sequence repeat and sequence-related amplified polymorphism markers. HortScience. 51(8):972–979.
- Zheng YZ, Xi JB, Yang ZY. 2005. Studies on distribution and morphological variation of wild grass Chrysopogon aciculatus collected from the tropics and subtropics of China. Acta Agrestia Sin. 3:117–122.