Abstract
Rubus lambertianus var. glaber is an excellent wild resource as a genetic donor. In this study, we sequenced the complete chloroplast genome of this species and evaluated its application in sequence diversity and phylogenetic analysis. The complete chloroplast genome exhibits a typical quadripartite structure, with two inverted repeats (257,790 bp each) separated by the large single-copy (LSC, 86,122 bp) and small single-copy (SSC, 18,867 bp) regions. A total of 133 genes were annotated, including 87 protein-coding genes, 38 tRNA, and 8 rRNA coding genes.Maximum-likelihood (ML) based phylogenetic analysis indicated that the R. lambertianus var. glaber has the closest relationship to Rubus fockeanus.
The Rubus genus of Rosaceae family contains hundreds of wide species and thousands of cultivated varieties worldwide (Hytönen et al. Citation2018). Raspberry and blackberry fruits are the typical known consumed brambles of this genus. Botanically, these plants are mainly from resources of subg. Rubus Idaeobatus, subg. Rubus malachobatus and subg. Rubus according to the classification system of Focke (Citation1914). Inter- and intra-species hybrids are also utilized in cultivation. However, in considering the occurrence of species (over 700) in the genus, vast majorities of resources remain unexplored. It is not until recently that the evolutionary history of Rubus species has been investigated using various molecular markers or high throughput sequencing techniques (Wang et al. Citation2016; Hummer et al. Citation2019). To date, only a limited number of chloroplast genome data are available for Rubus species.
Our previous investigation of the chromosomal affinity of wild Rubus species has identified the Rubus lambertianus var. glaber an excellent genetic donor for breeding (Wang et al. Citation2018). In this study, samples of this species were collected from Ya’an, Sichuan Province, China (29°58′24.5′′N, 103°00′18.7′′E). Voucher specimens were deposited in the herbarium of the College of Horticulture of the University with sample No. R01-8. DNA from fresh leaves was extracted using a modified CTAB protocol (Gawel and Jarret Citation1991). PCR amplifications especially targeting the chloroplast genome with 15 universal primer pairs referring to Zhang et al. (Citation2016) were carried out. PCR reaction components and reaction conditions were followed as in our previous report (Chen et al. Citation2019). After column purification, the equimolar of each PCR amplification product were pooled. Six micrograms of the DNA was used to construct 350 bp insertion libraries using the Illumina TruSeq DNA PCR-free library preparation protocol. Libraries were then sequenced on a HiSeq X Ten platform (BGI, Shenzhen, China).
To assemble the chloroplast genome, all pair-end short reads of 150 bp in length were first subjected to quality control using TrimGalore v0.6 (with parameters: q 20, length 15, and stringency 3) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Clean reads were then assembled using SPAdes v3.11 using kmer sizes of 23, 27, 31, 35, 39, 45, 55, 65, and 77 (Bankevich et al. Citation2012). The obtained contigs were mapped to the reference chloroplast genome of Fragaria chiloensis (Accession No. NC_019601) using Mummer 3.2 (Kurtz et al. Citation2004). Gaps in the genome were then filled up using a PCR based method. The complete chloroplast genome sequence has been deposed in the Genbank database under accession number MH992400.
To annotate the chloroplast genome, we used the GeSeq module with default settings (Tillich et al. Citation2017). The gene start and stop positions as well as intron and exon boundaries were manually curated. The R. lambertianus var. glaber chloroplast genome (156,569 bp) exhibited a typical quadripartite structure, with two IRs (257,790 bp, each) separated by the large single-copy (LSC, 86,122 bp) and small single-copy (SSC, 18,867 bp) regions. A total of 133 genes were annotated, including 87 protein-coding genes, 38 tRNA, and 8 rRNA coding genes.
With this genome sequence, we evaluated the chloroplast genome sequence divergence together with other public available Rubus chloroplast genome (Rubus corchorifolius, Rubus coreanus, Rubus crataegifolius, Rubus fockeanus, Rubus niveus, Rubus takesimensis and one from blackberry hybrid cultivar Arapahol). Genome sequences (with one repeat region being omitted) were aligned using progressive Mauve (Darling et al. Citation2004) with default parameters. Four large locally collinear blocks were identified with only small regions of rearrangement in the R. corchorifolus and R. coreanus. These aligned genome sequence were then investigated for sequence divergence using DNAsp v6 (Rozas et al. Citation2017). Nucleotide variability (Pi) within a sliding window (600 bp in width with step in size 200 bp) was evaluated. Sequences within genes trnQ-UUG, trnL-UUA, ndhF, ycf1, and trnL-UAG were found highly variable. Most of these gene loci were previously used in phylogenetic analysis.
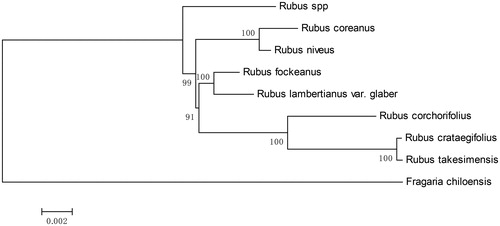
Phylogenomic analysis was then conducted with these sequences. Maximum-likelihood (ML) methods implemented in IQTREE v1.5.5 were used. Phylogenetic tree () revealed a closer relationship of R. lamberticanus var. glaber to the R. fockeanus than to the others, although they belong to different subgenera in the traditional taxon treatment system. This new chloroplast sequence provided valuable information for Rubus species diversity and phylogenetic applications in the future.
Figure 1. Maximum-likelihood (ML) tree based on conserved collinear blocks of the complete chloroplast genome of Rubus species, using Fragaria chiloensis as an outgroup. The numbers on the node are the fast bootstrap value based on 10,000 replicates. The bar indicates 0.002 mutations per site. Accession No. for the sequences used were listed as: Rubus spp. MH992399, Rubus coreanus MH992398, Rubus niveus KY419961, Rubus fockeanus KY420018, Rubus corchorifolius KY419958, Rubus crataegifolius MG189543, Rubus takesimensis NC_037991, Fragaria chiloensis NC_019601.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen Q, Wang Y, Sun B, Chen T, Luo Y, Zhang Y, Wang X-R, Tang H-R. 2019. The complete chloroplast genome sequence of Rubus coreanus, an excellent diseases-resistant resource. Mitochondrial DNA Part B. 4(1):216–217.
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Gen Res. 14(7):1394–1403.
- Focke WO. 1914. Species Ruborum: Monographiae generis Rubi Prodromus part III. New York (NY): Schweizerbart; p. 224–498.
- Gawel NJ, Jarret RL. 1991. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep. 9(3):262–266.
- Hummer KE, Carter KA, Liston A, Bassil NV, Alice LA, Bushakra JM, Sutherland BL, Mockler TC, Bryant DW. 2019. Target capture sequencing unravels Rubus evolution. Front Plant Sci. 10:1615.
- Hytönen T, Graham J, Harrison R. 2018. The genomes of Rosaceous berries and their wild relatives. New York (NY): Springer; p. 11–23.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Y, Chen Q, Chen T, Tang H, Liu L, Wang X. 2016. Phylogenetic insights into Chinese Rubus (Rosaceae) from multiple chloroplast and nuclear DNAs. Front Plant Sci. 7:968.
- Wang Y, Chen Q, Fu H, Zhang J, Chen T, Sun B, Luo Y, Zhang Y, Tang H, Wang X. 2018. Genome affinity in Rubus (Rosaceae) inferred from meiotic chromosome pairing of sixteen wild and cultivated bramble resources. Ind J Genet Plant Breed. 78(4):496–506.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J System Evol. 54(3):219–227.
