Abstract
Asobara japonica is an important larval-pupal endoparasitoid of Drosophila melanogaster and some other fruit fly species, such as Drosophila suzukii, and is an invasive and economic pest. In this study, the complete mitochondrial genome of A. japonica (GeneBank accession number: MN882556) was sequenced using Illumina HiSeq X Ten system. The mitochondrial genome is 15,519 bp long and comprises 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, of which 12 genes are in majority stand, and the remaining 25 genes are in minority strand. The nucleotide composition of A, G, C, T is 40.6, 8.2, 6.0, and 45.3% respectively. We also performed a phylogenetic analysis with other known mitochondrial genomes of Braconidae. The results show that A. japonica is closely related to Diachasmimorpha longicaudata.
Drosophila suzukii is a destructive pest of agriculture, especially damaging soft fruits such as berries, cherries, and wine grapes (Chen et al. Citation2018). Asobara japonica (Hymenoptera: Braconidae) is one of the most important natural enemies to constrain the D. suzukii population. Asobara japonica is a larval-pupal parasitoid, which can parasitize many species in Drosophila genus, and is thought to be an excellent biological control agent (Ideo et al. Citation2008). In order to increase the biological control efficacy, many studies have been done within the fields of Wolbachia-associated parthenogenesis and parasitic related behaviors of A. japonica, but few works have been done about their phylogenetic analysis and evolution (Kremer et al. Citation2009; Chabert et al. Citation2012; Furihata et al. Citation2016).
Asobara japonica was collected from Taizhou (28°50′N, 120°34′E), Zhejiang, China in June 2018. After sampling, the voucher specimen (ZJUHJH_001) was stored in 100% ethanol and kept in the Parasitic Hymenoptera Collection of Institute of Insect Sciences, Zhejiang University. The DNA of A. japonica was isolated and purified using the phenol–chloroform method. The complete mitochondrial genome of A. japonica (GeneBank accession number: MN882556) was sequenced using Illumina HiSeq X Ten system with the strategy of 150 paired-ends reading. It was further annotated using the Geneious 11.0.4 version and MITOS Web Server (Bernt et al. Citation2013).
The length of the complete mitochondrial genome of A. japonica is 15,519 bp, and it contains 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and a putative control region (CR). Further analysis revealed that 12 genes were encoded on the majority stand, and the remaining 25 genes were encoded on the minority strand. Three rearrangement events of tRNA clusters were found in the sequenced region similar to the putative ancestral arrangement of insects, corresponding to the I-M-Q, W-Y-C, and D-H-K patterns. The rearrangement pattern of tRNAs between cox2 and atp8 was D-H-K, which was consistent with previous results (Dowton Citation1999). Moreover, all of PCGs and tRNAs showed a similar order as the Pancrustacea Ancestral Gene Order (PanGo) (Wei et al. Citation2014). The overall base composition is 40.6% for A, 8.2% for G, 6.0% for C, and 45.3% for T, with an A + T content of 85.9%. Four start codons for PCGs were used: ATN (nad2, atp8, atp6, cox3, and nad5); ATT (nad6 and nad1); ATG (cox1 and nad4); ATA (cox2, nad3, nad4L, and cob), and all 13 PCGs used a TAA stop codon except for nad3, which ended with TAG. The 22 tRNAs genes varied from 64 to 69 bp in length, and the secondary structure of tRNAs was a typical clover-leaf structure as with other insects. The rrnL was located between trnL1 and trnV, whereas rrnS was between trnV and trnI. The length of rrnL and rrnS was 1366 and 751 bp, respectively.
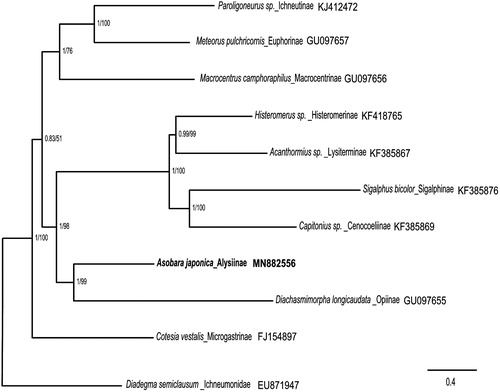
The complete mitochondrial genome of the genus Asobara is limited. Thus, we performed the phylogenetic analysis of A. japonica with 11 other insects in family Braconidae based on nucleotide sequences of the 13 PCGs and two rRNAs from their mitochondrial genomes (Li et al. Citation2016). The sequences were aligned by using MAFFT v7.271, and the phylogenetic tree was constructed by CIPRES (https://www.phylo.org/) using RAxML-HPC2 on XSEDE with bootstrap 1000 and MrBayes on XSEDE (). Phylogenetic analysis showed that A. japonica is closely related to Diachasmimorpha longicaudata. This study would further clarify our understanding of the phylogenetic relationship of the Braconidae family.
Figure 1. Phylogenetic relationships among subfamilies of the Braconidae inferred from nucleotides of 13 PCGs and two rRNAs using Bayesian and maximum-likelihood (ML) methods (GenBank accession numbers provided). The Bayesian posterior probabilities (PP) and bootstrap support (BS) are marked besides the nodes. Diadegma semiclausum was set as the outgroup.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chabert S, Allemand R, Poyet M, Eslin P, Gibert P. 2012. Ability of European parasitoids (Hymenoptera) to control a new invasive Asiatic pest, Drosophila suzukii. Biol Control. 63(1):40–47.
- Chen J, Zhou S, Wang Y, Shi M, Chen X, Huang J. 2018. Biocontrol characteristics of the fruit fly pupal parasitoid Trichopria drosophilae (Hymenoptera: Diapriidae) emerging from different hosts. Sci Rep. 8(1):13323.
- Dowton M. 1999. Relationships among the cyclostome braconid (Hymenoptera: Braconidae) subfamilies inferred from a mitochondrial tRNA gene rearrangement. Mol Phylogenet Evol. 11(2):283–287.
- Furihata S, Matsumura T, Hirata M, Mizutani T, Nagata N, Kataoka M, Katayama Y, Omatsu T, Matsumoto H, Hayakawa Y. 2016. Characterization of venom and oviduct components of parasitoid wasp Asobara japonica. PLoS One. 11(7):e0160210.
- Ideo S, Watada M, Mitsui H, Kimura MT. 2008. Host range of Asobara japonica (Hymenoptera: Braconidae), a larval parasitoid of drosophilid flies. Entomol Science. 11(1):1–6.
- Kremer N, Charif D, Henri H, Bataille M, Prévost G, Kraaijeveld K, Vavre F. 2009. A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity. 103(3):248–256.
- Li Q, Wei S, Tang P, Wu Q, Shi M, Sharkey MJ, Chen X. 2016. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol Evol. 8(9):2651–2662.
- Wei S, Li Q, Van Achterberg K, Chen X. 2014. Two mitochondrial genomes from the families Bethylidae and Mutillidae: independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol Phylogenet Evol. 77:1–10.
