Abstract
Species in the genus Botrychium are tiny herbaceous plants with simple morphology that are widely distributed in the northern hemisphere. The phylogenetic relationship intra the genus is not well resolved, different species/populations from North America, Europe, and Asia demonstrate a paraphyletic relationship, including Botrychium lunaria. In this study, we reported the first complete chloroplast genome sequence of B. lunaria from Chinese population. It is 138,558 bp in length, and composed with a typical quadripartite structure. The overall GC content is 44.40%. In total, 129 genes were annotated, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A maximum-likelihood tree of primitive fern lineage based on complete chloroplast genome sequence data was constructed. The phylogenetic analysis indicated that Botrychium is closely related to other basal fern lineages, such as Helminthostachys, Ophioglossum, and Mankyua. Furthermore, the genomic data generated in this study will provide helpful information for the conservation of B. lunaria in Beijing, China.
The genus Botrychium, commonly called moonwort, belongs to the basal clade of fern lineage Ophioglossaceae with 35 accepted species and 13 uncertain taxa (Dauphin et al. Citation2017). This genus is broadly distributed in cool temperate and boreal regions of Asia, Australia, Europe, New Zealand, North America, and temperate South America (Farrar and Stensvold Citation2017; Meza-Torres et al. Citation2017). Because of the tiny size (range from 1 to 15 cm in height) and a few distinguishing morphological characters, species in Botrychium are difficult to differentiate and many cryptic ones exist, such as the Botrychium lunaria complex. Samples of B. lunaria from North America, Europe, and Asia are not resolved as a monophyletic clade, which implies the complicated genetic background of B. lunaria (Dauphin et al. Citation2017, Citation2018; Maccagni et al. Citation2017; Stensvold and Farrar Citation2017). Botrychium lunaria is listed in the first-level key protected wild plants in Beijing City. A deep genomic investigation will facilitate the conservation of this rare species in Beijing. Hence, we reassembled and annotated the complete chloroplast genome sequence of B. lunaria of the Asian population with the purpose to provide more genetic information of this complicated species and facilitate further phylogenetic research of the genus Botrychium.
The fresh leaves of B. lunaria were collected from Song Mountain in Yanqing District, Beijing, China (N40°31′10.81′′, E115°49′14.95′′). The voucher specimen (collector and collection number: Xian-Yun Mu and 4449) is stored in the herbarium of Beijing Forestry University (BJFC). The extracted genomic DNA was sequenced by the next-generation method on Illumina Hiseq platform. In total, 2.77 GB of 150-bp clean reads were assembled into the complete chloroplast genome using Geneious Prime 2020.0.4 software (Biomatters Ltd., Auckland, New Zealand) with Botyhcium ternatum/japonicum, Helminthostachys zeylanica, Mankyua chejuensis, Ophioglossum californicum as references. The genome was annotated using PGA (Qu et al. Citation2019), adjusted manually on Geneious Prime 2020.0.4, and deposited in NCBI with the accession number MN966674.
The complete chloroplast genome of B. lunaria was 138,558 bp long with the typical quadripartite structure consisting of a pair of inverted repeat regions (IRs with 9,474 bp), and two single-copy regions (LSC with 99,013 bp; SSC with 20,597 bp). Overall, the GC content in whole genome, LSC, SSC, and IR is 44.40, 43.19, 42.35, and 52.64% respectively. The genome contains 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
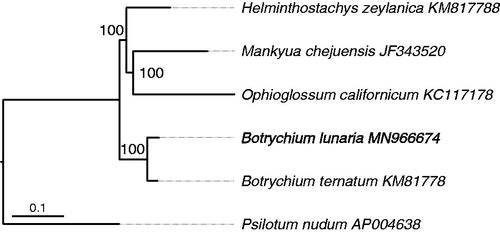
A maximum-likelihood (ML) tree was constructed with the complete chloroplast genomes of six ferns representing five genera. Sequences were aligned in MAFFT online service (Katoh et al. Citation2017), and then the poorly aligned positions and divergent regions were eliminated by GBLOCKS 0.91b (Talavera and Castresana Citation2007). Finally, the phylogenetic tree was generated using IQ-TREE (Hoang et al. Citation2018) with 1000 bootstrap replicates (). The obtained ML tree is well resolved, and the generic topology is consistent with previous studies (Hauk et al. Citation2003; Shen et al. Citation2018). The two moonwort species are clustered into a clade and come together with others as a whole, and the genus Botrychium exhibited a close relationship with other relatively primitive fern lineages, such as Helminthostachys, Ophioglossum, and Mankyua. Our empirical data obtained a robust phylogeny of the basal clade of fern lineages, and it is worthy to investigate the phylogeny and biogeography of Botrychium by using the complete chloroplast genome sequence data in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dauphin B, Farrar DR, Maccagni A, Grant JR. 2017. A worldwide molecular phylogeny provides new insight on cryptic diversity within the moonworts (Botrychium s.s., Ophioglossaceae). Syst Bot. 42(4):620–639.
- Dauphin B, Grant JR, Farrar DR, Rothfels CJ. 2018. Rapid allopolyploid radiation of moonwort ferns (Botrychium; Ophioglossaceae) revealed by PacBio sequencing of homologous and homeologous nuclear regions. Mol Phylogenet Evol. 120:342–353.
- Farrar DR, Stensvold MC. 2017. Observations on bipolar disjunctions of moonwort ferns (Botrychium, Ophioglossaceae)). Am J Bot. 104(11):1675–1679.
- Hauk WD, Parks CR, Chase MW. 2003. Phylogenetic studies of Ophioglossaceae: evidence from rbcl and trnl-F plastid DNA sequences and morphology. Mol Phylogenet Evol. 28(1):131–151.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Maccagni A, Parisod C, Grant JR. 2017. Phylogeography of the moonwort fern Botrychium Lunaria (Ophioglossaceae) based on chloroplast DNA in the Central-European mountain system. Alp Botany. 127(2):185–196.
- Meza-Torres EI, Stensvold M, Farrar D, Ferrucci M. 2017. Circumscription of the South American moonwort Botrychium (Ophioglossaceae). Plant Biosyst. 151(2):258–268.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Shen H, Jin DM, Shu JP, Zhou XL, Ming L, Wei R, Shang H, Wei HJ, Zhang R, Liu L, et al. 2018. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience. 7(2):gix116.
- Stensvold MC, Farrar DR. 2017. Genetic diversity in the worldwide Botrychium Lunaria (Ophioglossaceae) complex, with new species and new combinations. Brittonia. 69(2):148–175.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.