Abstract
Dendrocalamus brandisii is high bamboo with branches several, dominant branches well developed. In this study, the complete plastid genome of D. brandisii was assembled and annotated. The total plastid genome was 139,423 bp in length, consisting a large single-copy region (LSC) of 82,956 bp, a small single-copy region (SSC) of 12,879 bp, and a pair of inverted repeats (IR) regions of 21,794 bp. The complete chloroplast genome contains 132 genes, including 39 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes and 85 protein-coding genes. The phylogenetic analysis suggested that D. brandisii sister to D. latiflorus.
Woody bamboos (Poaceae: Bambusoideae) comprise ca. 80 genera including more than 1200 species, which are mainly distributed in the subtropical and tropical regions of Asia, America and Africa (Li et al. Citation2006; Yi et al. Citation2008). Dendrocalamus is a woody bamboo genus placed in the subtribe Bambusinae and tribe Bambuseae (Ohrnberger Citation1999). Species of the mentioned genera are characterized by their sympodial rhizomes and large-sized, dense clumps and it contains over 50 species, naturally distributed in tropical and subtropical regions of the Old World (Pattanaik and Hall Citation2011).
Fresh Leaf sample of D. brandisii was acquired from Cangshan district, Fuzhou city, Fujian Province of China (26°38′N, 119°27′E), and the voucher specimen deposited at the Herbarium of Fujian Agriculture and Forestry University (specimen code HTY004). The total genomic DNA was extracted from fresh leaves using a modified Cetyltrimethylammonium Ammonium Bromide (CTAB) method (Doyle and Doyle Citation1987) and sequenced by the BGISEQ-500 platform. The samples were collected from clean reads were used to assemble the complete chloroplast genome by the GetOrganelle pipe-line (Jin et al. Citation2018), with the chloroplast genome of Dendrocalamus latiflorus (GenBank accession NC_013088) as the reference sequences. The assembled chloroplast genome was annotated using the Geneious R11.15 (Kearse et al. Citation2012).
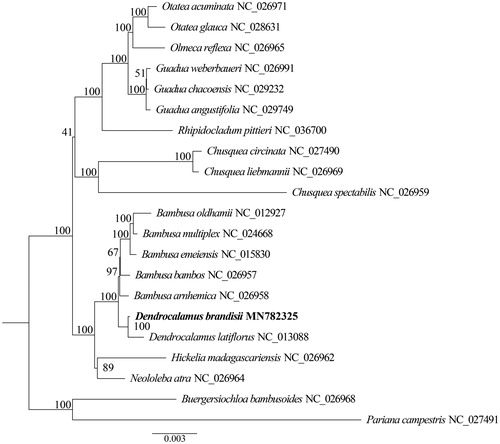
The complete plastid genome sequence of D. brandisii (GenBank accession MN782325) was 139,423 bp in length, consisting of a large single-copy region (LSC) of 82,956 bp, a small single-copy region (SSC) of 12,879 bp, and a pair of inverted repeats (IR) regions of 21,794 bp. The GC content of the chloroplast genome was 38.91%. Complete plastid genome contains 132 genes, including 39 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes and 85 protein-coding genes. To confirm the phylogenetic position of D. brandisii, a phylogenetic analysis was performed based on 20 representative plastid genome sequences. All sequences were aligned with the HomBlock pipeline (Bi et al. Citation2018), and the phylogenetic tree constructed by RAxML (Stamatakis Citation2014). The ML tree showed that D. brandisii is sister to D. latiflorus with strong support. ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton C. 2006. Bambuseae (Poaceae). Flora of China. vol. 22. Beijing: Science Press; p. 1.
- Ohrnberger D. 1999. The bamboos of the world: annotated nomenclature and literature of the species and the higher and lower taxa. Amsterdam: Elsevier; p. 585.
- Pattanaik S, Hall JB. 2011. Molecular evidence for polyphyly in the woody bamboo genus Dendrocalamus (subtribe Bambusinae). Plant Syst Evol. 291(1–2):59–67.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yi TP, Shi JY, Ma LS, Wang HT, Yang L. 2008. Iconographia Bambusoidearum sinicarum. Beijing: Science Press; p. 766.