Abstract
We describe the mitogenome sequence of Euproctis seitzi collected in the Mt. Luofu, which located in the southeast of China. The assembled mitogenome is 15, 276 bp in length and consists of 13 protein-coding genes, 22 transfer-RNA genes, 2 ribosomal-RNA genes, and one A + T rich region. The most common start codon for 13 PCGs is ATT and the most common termination codon is TAA. The overall G + C content was only 18.57% in the heavy strand. The result of the phylogenetic analysis shows that the relationship of E. seitzi is close to the species in the same subfamily.
Euproctis seitzi Stand, 1910 (Lepidoptera: Erebidae, Lymantriinae) is a tussock moth mainly distributed in the southern coastal regions of China, which was first described by Strand (Citation1910) from Hongkong. Different with other species in the same genus, such as E. pseudoconspersa which is a notorious pest mainly feeding on the leaves of tea trees (Dong et al., Citation2016), E. seitzi is a typically euryphagous insects. It could cause enormous economic loss in economic trees, such as oranges and mulberries (Chao, Citation1987, Citation2003). Moreover, the hairy caterpillars of E. seitzi might cause outbreaks of irritating cutaneous eruptions due to the venomous spicules on their backs (Bleumink et al. Citation1982; de Jone et al. Citation1982). Although previous studies on the genus Euproctis have mainly focused on the identification based on morphological characteristics of adults (Chao, Citation1984, Citation1986; Wang et al., Citation2010), there were still challenges in species identification due to the short-term emergence of the adults (mainly in June to August). Other studies of Euproctis focused on sex pheromone synthesis (Leonhardt et al. Citation1991; Wakamura et al. Citation1994), or the nuclear polyhedrosis virus that infects larvae (Kawamoto et al. Citation1977; Tang et al. Citation2009). Molecular information, especially mitogenome which can provide systematically-informative information for identification and phylogenetic analysis, are under development.
We collected the samples of E. seitzi in Mt. Luofu, Guangdong Province, China (23°17.5′N and 114°12.3′E) in July 2019. All specimens were preserved in 95% ethanol in the field and stored at 4 °C in the laboratory until DNA extraction. Voucher specimen was deposited at the Entomological Museum of Capital Normal University (No. VLym-19.2016) and the sequence was deposited in GenBank (No. MN916588).
The length of the complete mitochondrial genome of E. seitzi is 15,276bp, which contains 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 control region (A + T-rich region) which is 461 bp in length. For the 13 PCGs, the most common start codon is ATT (ATP8, COX2, ND1, ND2, ND3, ND6), then is ATG (ATP6, COX3, Cytb and ND4) and ATA (COX1, ND4L and ND5); the most common termination codon is TAA (13 protein coding genes except ND4L). The mitochondrial base composition is A 39.55%, T 41.88%, G 7.08%, and C 11.49% in the heavy strand, with an obvious (A + T)% > (G + C)%. Similar situation occurred in the non-coding region in which the (A + T) % was more than 90%. The length of the 22 sequenced tRNA genes ranged from 61 to 71 bp. The lrRNA is 1272 bp long with an A + T content of 84.33% and the srRNA is 811 bp long with an A + T content of 87.54%.
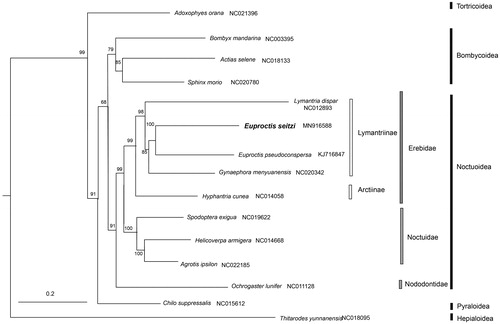
All 13 protein-coding genes were used to reconstruct the phylogenetic tree with the maximum Likelihood approach in RAxML 7.9.6 (Stamatakis Citation2006). The two Euproctis species were clustered into a branch with 100 bootstrap values. Meanwhile, the analysis confirmed the monophyly of the superfamily Noctuoidea (including Noctuidae, Erebidae and Notodontidae) ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bleumink E, de Jong MCJM, Kawamoto F, Meyer GT, Kloosterhuis AJ, Slijper-Pal IJ. 1982. Protease activities in the spicule venom of Euproctis caterpillars. Toxicon. 20(3):607–613.
- Chao CL. 1984. New species of the genus Euproctis Hűbner from China (Lepidoptera: Lymantriidae). Acta Entomol Sin. 27(4):449–456.
- Chao CL. 1986. Two new species of the Chinese Euproctis (Lepidoptera: Lymantriidae). Acta Entomol Sin. 29(4):432–433.
- Chao CL. 1987. Economic insect fauna of China (in Chinese). Volume 42. Beijing: Science Press.
- Chao CL. 2003. Fauna sinica insect (in Chinese). Volume 30. Beijing: Science Press.
- Dong WW, Dong SY, Jiang GF, Huang GH. 2016. Characterization of the complete mitochondrial genome of tea tussock moth, Euproctis pseudoconspersa (Lepidoptera: Lymantriidae) and its phylogenetic implications. Gene. 577(1):37–46.
- de Jone M, Kawamoto F, Bleumink E, Kloosterhuis AJ, Meijer GT. 1982. A comparative study of the spicule venom of Euproctis caterpillars. Toxicon. 20:477–485.
- Kawamoto F, Kumada N, Kobayashi M. 1977. Envelopment of the nuclear polyhedrosis virus of the oriental tussock moth, Euproctis subflava. Virology. 77(2):867–871.
- Leonhardt BA, Mastro VC, Schwarz M, Tang JD, Charlton RE, Pellegrini-Toole A, Warthen JD, Schwalbe CP, Cardé RT. 1991. Identification of sex pheromone of browntail moth, Euproctis chrysorrhoea (L.) (Lepidoptera: Lymantriidae). J Chem Ecol. 17(5):897–910.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Strand E. 1910. Lymantriidae. in Seitz. Gross-Schmett. Erde. 2:109–141.
- Tang XD, Xiao Q, Ma XC, Zhu ZR, Zhang CX. 2009. Morphology and genome of Euproctis pseudoconspersa nucleopolyhedrovirus. Virus Genes. 38(3):495–506.
- Wakamura S, Yasuda T, Ichikawa A, Fukumoto T, Mochizuki F. 1994. Sex attractant pheromone of the tea tussock moth, Euproctis Pseudoconspersa (Strand) (Lepidoptera: Lymantriidae): identification and field attraction. Appl Entomol Zool. 29(3):403–411.
- Wang HS, Fan XL, Wang M. 2010. A new record of Euproctis wilemani (Lepidoptera: Lymantriidae) from Hainan Island. Florida Entomol. 93(2):325–326.