Abstract
Palmaria decipiens (Reinsch) R.W.Ricker is a ecologically important red seaweed restricted to high latitudes of the southern hemisphere. Here, we contribute to the bioinformatics and evolutionary systematics of the Palmariales by performing high throughput sequencing analysis on a specimen of P. decipiens from the western Antarctic Peninsula. The P. decipiens mitogenome is 26,645 base pairs (bp) in length and contains 49 genes (GenBank accession MN967053) and the plastid genome is 193,007 bp and contains 245 genes (GenBank accession MN967052). The mitogenome and plastid genome of P. decipiens are similar to P. palmata from Japan in pairwise genetic distances (93.71% and 98.14%, respectively), and P. palmata from the Maine, USA (87.45% and 94.57%, respectively). The genomes of P. decipiens showed high gene synteny with P. palmata, however several tRNA differences are documented. Organellar genome content and phylogenetic analyses of P. decipiens supports its placement in the genus Palmaria.
Palmaria decipiens is a dominant marine red algal ecosystem species that occurs in the intertidal and subtidal, where it provides habitat, nourishment and shelter for many marine organisms (Becker et al. Citation2011). It is characterized as having reddish to purple, unbranched blades, that extend up to 70 cm long and have a lubricous glossy surface (Ricker Citation1987; Becker et al. Citation2011). To better understand the taxonomy of Palmaria and P. decipiens, the complete mitogenome and plastid genome of P. decipiens from Yelcho Chilean station, Doumer Island, Antarctic Peninsula (64°52′41″S, 63°35′51″W) were characterized.
DNA was extracted from P. decipiens (Specimen Voucher- LMS000004 in HIP, see Thiers Citation2016) using the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. The 150 bp PE Illumina library construction and sequencing was performed by myGenomics, LLC (Alpharetta, Georgia, USA). The genomes were assembled using default de novo settings in MEGAHIT (Li et al. Citation2016) and Geneious Prime to close the gaps (Biomatters, Ltd, Auckland, New Zealand). The genes were annotated manually using blastx, NCBI ORFfinder, tRNAscan-SE 1.21 (Schattner et al. Citation2005), and RNammer (Lagesen et al. Citation2007). The P. decipiens plastid genome was aligned to other plastomes using MAFFT (Katoh and Standley Citation2013). The phylogenetic analysis was executed with RAxML-NG (Kozlov et al. Citation2019) using the GTR + gamma model and 1000 bootstraps. The tree was visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
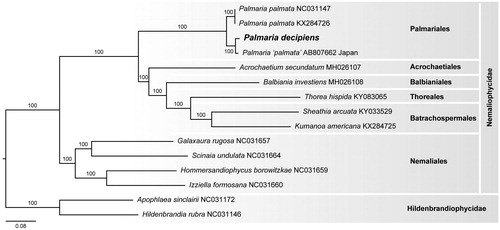
The mitogenome of P. decipiens is 26,645 bp in length and contains 49 genes. It is A + T-rich (67.0%) and includes 23 tRNA, 5 ribosomal proteins, 2 rRNA (rrl, rrs), and 19 other genes involved in mitochondrial function. The plastid genome of P. decipiens is 193,007 bp and contains 245 genes. It is also A + T biased (65.2%) and includes 45 ribosomal proteins, 33 tRNA, 32 photosystem I and II, 30 ycf, 12 cytochrome b/f complex, 8 ATP synthase, 4 RNA polymerase, 6 rRNA, and 75 other genes. The mitogenome of P. decipiens lacks the trnI gene, but like P. palmata from Japan contains trnD and trnH (Kumagai et al. Citation2019). The plastid genome of P. decipiens differs from P. palmata from Japan and Maine, USA, by the addition of the trnE gene (Costa et al. Citation2016; Kumagai et al. Citation2019). Pairwise genetic distances of the mitogenome and plastid genome of P. decipiens and P. palmata from Japan are more similar in mitogenome (93.71%) and plastid genome (98.14%) sequence than P. decipiens is to P. palmata from the Maine, USA (87.45% and 94.57%, respectively). These organellar data support the existence of three distinct entities: P. palmata from the Atlantic; P. ‘palmata’ from Japan; and P. decipiens from Antarctica.
Phylogenetic analysis of the plastid genome of P. decipiens positions it in a clade with P. palmata from Japan and the USA (). This evolutionary relationship is similar to the most recent analyses in which the Palmariales is closely allied with the Acrochaetiales, Balbianiales, and Nemaliales (Costa et al. Citation2016; Yang et al. Citation2016; Saunders et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Becker S, Quartin ML, Campana GL, Bucolo P, Wiencke C, Bischof K. 2011. The biology of an Antarctic rhodophyte, Palmaria decipiens: recent advances. Antartic Sci. 23(5):419–430.
- Costa JF, Lin SM, Macaya EC, Fernandez-Garcıa C, Verbruggen H. 2016. Chloroplast genomes as a tool to resolve red algal phylogenies: a case study in the Nemaliales. BMC Evol Biol. 16:205.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server):W465–W469.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Kumagai Y, Tsubouchi R, Miyabe Y, Takeda T, Adachi K, Yasui H, Kishimura H. 2019. Complete sequence of mitochondrial DNA of red alga dulse Palmaria palmata (Linnaeus) Weber & Mohr in Japan. Mitochondrial DNA Part B. 4:3177–3178.
- Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. 2007. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 35(9):3100–3108.
- Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102:3–11.
- Ricker RW. 1987. Taxonomy and biogeography of Macquarie Island seaweeds. London: British Museum Press.
- Saunders GW, Jackson C, Salomaki ED. 2018. Phylogenetic analyses of transcriptome data resolve familial assignments for genera of the red-algal Acrochaetiales-Palmariales complex (Nemaliophycidae). Mol Phylogenet Evol. 119:151–159.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:686–689.
- Thiers B. 2016. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. [accessed 2020 Feb 24]. http://sweetgum.nybg.org/science/ih/.
- Yang EC, Boo SM, Bhattacharya D, Saunders GW, Knoll AH, Fredericq S, Graf L, Yoon HS. 2016. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep. 6(1):21361.