Abstract
The complete mitochondrial genome of Rhytidodus viridiflavus was determined for the first time in the present study. Based on annotation, the mitogenome was found to have circular shape, with 16,842 bp in size, containing 13 protein-coding genes (PCGs), 22 transfer-RNA genes, 2 ribosomal-RNA genes, and 1 non-coding region. All PCGs have ATN as the start codon, TAA and single T— as the stop codon. The concatenated PCGs were used to perform Bayesian phylogenetic analyses together with mitogenome data of Hemiptera in GenBank. The resulting phylogenetic tree confirms that the R. viridiflavus belongs to the subfamily of Idiocerinae, being closely related to Populicerus popili plus Idiocerus laurifoliae. The mitogenome reported herein will provide useful genetic markers for studying the molecular ecology, systematics, and population genetics of Idiocerine leafhoppers.
Leafhopper is one of the most widely distributed and abundant group in agroforestry ecosystems, which comprises ∼22,000 species all over the world (Camisão et al. Citation2014). The species of Idiocerinae are widespread throughout all zoogeographic regions, with approximately 106 genera and 800 species recorded (Zhang and Webb Citation2019). However, our understanding of the mitochondrial genomes of Idiocerinae, even of all leafhoppers, is very limited (Choudhary et al. Citation2018; Wang et al. Citation2018).
In this study, we sequenced and annotated the complete mitochondrial genome (mitogenome) of R. viridiflavus using next-generation sequencing through the Illumina HiSeq 2000 platform. A male adult was selected as a specimen (voucher number IMNU20190706), which was collected from the type locality of Shawan County, Xinjiang Uygur Autonomous Region, China, in July 2019. The entire body without abdomen was shipped to Tsingke (Beijing, China) for genomic extraction and 150-base-pair paired-end library construction; sequencing was performed on an Illumina HiSeq 2000 instrument. De novo assembly of clean reads was performed using SPAdes v3.11.0 (Bankevich et al. Citation2012). The mitogenome of Idioscopus clypealis (GenBank accession number NC_039642) was further used as a reference to assemble the sequenced sample. Genes were annotated with the MITOS (Bernt et al. Citation2013) web server. The annotated sequence of R. viridiflavus mitogenome was deposited in GenBank with an accession number MN935488.
The mitochondrial genome of R. viridiflavus is 16,842bp in length, with the A + T content of 78% (T 34.1%, C 13.0%, A 43.9%, and G 9.0%), which is within the range reported for hemipteran mitogenomes (68.86–86.33%; Zhang et al. Citation2014). Annotation of the mitogenome revealed 13 protein-coding genes (PCGs) (COX1-3, ND1-6, ND4L, ATP6, ATP8, and Cytb), 22 transfer-RNA (tRNA) genes, and 1 control region (CR or D-loop). In 13 PCGs, only four genes (ND4, ND4L, ND5, and ND1) are coded on the L-strand, whereas the others are coded on the H-strand. The total length of 13 PCGs is 10,939 bp, which have ATN as the start coding, TAA and a single T— as the stop codon. The concatenated PCGs were used to perform Bayesian phylogenetic analyses together with mitogenome data of Hemiptera in GenBank; sequences were aligned using MEGA6 software (Tamura et al. Citation2013). All tRNA genes were identified by tRNAscan-SE Search Server (Lowe and Chan Citation2016). The 16S rRNA gene (640 bp) is located between tRNA-Leu1 and tRNA-Val, whereas the 12S rRNA gene (751 bp) is located after tRNA-Val. The D-loop is 2503 bp in size.
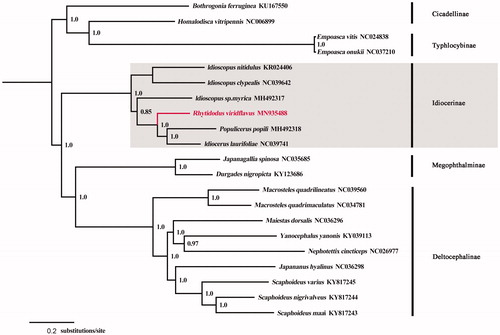
Phylogenetic analyses of R. viridiflavus and other 20 Hemiptera taxa were conducted based on the concatenated PCGs using Bayesian inference (BI) method in MrBayes v3.2.1 (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012) under the GTR + G model. The resulting phylogenetic tree () shows that R. viridiflavus is nested within the subfamily Idiocerinae, being closely related to Populicerus popili plus Idiocerus laurifoliae. The mitogenome will provide useful genetic markers for studying the molecular ecology, systematics, and population genetics of idiocerine leafhoppers.
Figure 1. A majority-rule consensus tree (midpoint rooted) inferred from Bayesian inference using MrBayes v3.2.1 under the GTR + G model, based on the concatenated PCGs. Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution. GenBank accession numbers are given with species names.
Acknowledgements
We thank Dr. Xianguang Guo (Chengdu Institute of Biology, Chinese Academy of Sciences) for his assistance in analyzing the data and revising the manuscript. We also thank Ms. Minli Chen for her helpful suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69(2):313–319.
- Camisão BM, Cavichioli RR, Takiya DM. 2014. Eight new species of Oragua Melichar, 1926 (Insecta: Hemiptera: Cicadellidae) from Amazonas State, Brazil, with description of the female terminalia of Oragua jurua Young, 1977, and new records for the genus. Zootaxa. 3841(4):501–527.
- Choudhary JS, Naaz N, Das B, Bhatt BP, Prabhakar CS. 2018. Complete mitochondrial genome of Idioscopus nitidulus (Hemiptera: Cicadellidae). Mitochondrial DNA B. 3:32–33.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang JJ, Yang MF, Dai RH, Li H, Wang XY. 2018. Characterization and phylogenetic implications of the complete mitochondrial genome of Idiocerinae (Hemiptera: Cicadellidae). Int J Biol Macromol. 120:2366–2372.
- Zhang B, Webb MD. 2019. A checklist and key to the idiocerine leafhoppers (Hemiptera: Cicadellidae) of Hainan Island, with description of a new genus and new species. Zootaxa. 4576(3):581–587.
- Zhang KJ, Zhu WC, Rong X, Liu J, Ding XL, Hong XY. 2014. The complete mitochondrial genome sequence of Sogatella furcifera (Horváth) and a comparative mitogenomic analysis of three predominant rice planthoppers. Gene. 533(1):100–109.
