Abstract
Acorales comprises the basal lineage of all extant monocots and has important linkage to early angiosperms and dicots. In this study, we reported the complete chloroplast genome of Acorus calamus, one of the two species of Acorales. The whole genome was 152,876 bp in length, consisting of a pair of inverted repeats (IR 25,661 bp), a large single-copy region (LSC 83,290 bp), and a small single-copy region (SSC 18,264 bp). The complete genome contained 129 genes, including 84 protein-coding genes, 37 tRNA, and 8 rRNA genes. The overall GC content of the whole genome was 38.7%. A maximum-likelihood phylogenetic analysis showed that A. calamus is sister to A. calamus which was collected in Germany. The complete chloroplast genome of A. calamus will help improve and integrate the existing genome data of monocots and provide insights into the phylogenetic relationship among basal angiosperms, monocots and dicots.
Identifying the phylogenetic relationship among the major lines of angiosperms has long been a subject of debate. As the oldest lineage of monocotyledon, Acorales only has a family Acoraceae, a genus Acorus and two species (Govaerts and Frodin Citation2002; Givnish et al. Citation2018), which makes it an important position for revealing the evolution and diversification of monocots. One of the two species, Acorus calamus, commonly known as sweet flag, is an herbaceous perennial that distributes throughout aquatic habitats such as water edge, swamp, wetlands at elevation below 2600 meters in temperate and subtropical regions over the world (Motley Citation1994; Govaerts and Frodin Citation2002; Balakumbahan et al. Citation2010). Previously, a chloroplast genome of A. calamus (Goremykin et al. Citation2005) collected at botanical garden of the University of Jena, Germany has been reported. To compare chloroplast of A. calamus from different distributed regions, we sequenced complete chloroplast genome of A. calamus colleted from Fujian, China.
Leaf sample of A. calamus was collected from Meihua Mountain, Fujian Province, China (25°15′29.67″N, 116°45′20.83″E). The DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequencing was carried out by the Illumina Hiseq Xten platform, with approximately 67.3 GB data generated. The DNA was stored at Fujian Agriculture and Forestry University (Voucher specimen: AC-01, FAFU). The clean reads were aligned to A. calamus published by Goremykin et al. (Citation2005) (GenBank accession No. NC_007407) and assembled by GetOrganelle pipe-line (Jin et al. Citation2018) to obtain plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. Citation2015). The assembled chloroplast genome was annotated using DOGMA (Wyman et al. Citation2004), and corrected by Geneious 11.1.5 (Kearse et al. Citation2012) using A. calamus (NC_007407) as reference sequences. The physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013). The annotated complete chloroplast genome was submitted to GenBank with accession number MN9670114.
The complete chloroplast genome of A. calamus is 152,876 bp in length, comprising a large single-copy (LSC) region of 83,290 bp, a small single-copy (SSC) region of 18,264 bp, and two inverted repeat (IR) regions of 25,661 bp. The new sequence contained 129 genes in total, including 84 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The base composition of A. calamus cp genome was uneven (30.3% A, 19.8% C, 19.0% G, 30.9% T) with an overall GC content of 38.7% and the corresponding values of the LSC, SSC, and IR regions reaching 37.4%, 33.2%, and 42.9%, respectively. The result was different from A. calamus (NC_007407) collected in Germany (Goremykin et al. Citation2005), which showed a cpDNA length of 153,821 and 130 genes in total, indicating different annotation methods and population variations of same species could yield different results in chloroplast genomes.
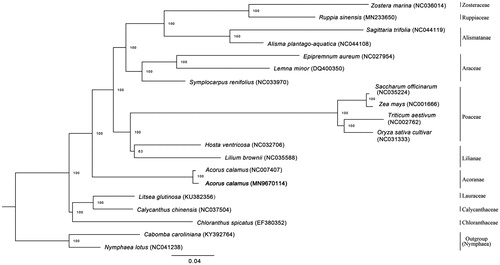
To investigate the phylogenetic position of A. calamus, 20 complete cp genomes of species from Lauraceae, Calycanthaceae, Chloranthaceae, Acoranae Alismatanae, Araceae, Ruppiaceae Zosteraceae, Lilianae and Poaceae, downloaded from NCBI, were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with Camboba caroliniana and Nymphaea lotus as outgroup. The branch support was computed with 1000 bootstrap replicates. The ML tree analysis indicated that A. calamus and A. calamus (NC_007407) cluster together with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Balakumbahan R, Rajamani K, Kumanan K. 2010. Acorus calamus: an overview. J Med Plant Res. 4:2740–2745.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Givnish T J, Zuluaga A, Spalink D, Soto Gomez M, Lam V K. Y, Saarela J M, Sass C, Iles W J. D, de Sousa D J L, Leebens-Mack J, et al. 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi‐gene analyses, and a functional model for the origin of monocots. Am J Bot. 105(11):1888–1910.
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. 2005. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 22(9):1813–1822.
- Govaerts R, Frodin DG. 2002. World checklist and bibliography of Araceae. Kew: Royal Botanical Gardens.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. doi:10.1101/256479
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Motley TJ. 1994. The ethnobotany of sweet flag, Acorus calamus (Araceae). Econ Bot. 48(4):397–412.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.