Abstract
Epimedium dewuense S. Z. He is a rare and endangered endemic to China. The complete chloroplast (cp) genome of E. dewuense was reported in this study. The total cp genomes size was 157,056 bp in length, with 38.8% of GC content, and a typical quadripartite structure, the large single-copy region (LSC, 88,346 bp), small single-copy region (SSC, 26,768 bp) that separate by a pair of inverted repeat regions (IRs, each for 20,820 bp). The whole cp genome of E. dewuense contained 130 unique genes, including 37 tRNA, 7 rRNA, and 86 protein-coding genes. This plastid genome is the first report for the E. dewuense and will be useful data for developing markers for further studies on resolving the relationship within the Berberidaceae.
Epimdium belongs to Beiberdaceae, up to now, there are 68 species in the world, and about 59 species distribution in China, (Ying et al. Citation2000; He Citation2014; Zhang et al. Citation2016). Epimdium is a valuable plant resource, and famous for its medicinal. Epimedii folium has the therapeutic effects of nourishing kidney, strengthening bones and reliev-ing rheumatism, and has a long history of medical use intraditional Chinese medicine (Wu et al. Citation2010; Ma et al. Citation2011; Jiang et al. Citation2016). E. dewuense is a new species,and was discovered by S. Z. He in Citation2003, which is a rare and endangered endemic to China and only distributed at the junction of Wuchuan and Dejiang counties in Guizhou, China, with a very small population (He and Xu Citation2003). In this study, we sequenced the complete cp genomes of E. dewuense, aiming to provide valuable information for studying resource protection tax-onomy and phylogeny of Epimedium genus.
The E. dewuense samples in this study were collected in the Fengle town, Wuchuan county, Guizhou province, China, (N 28°20′32.79″, E 107°51′56.44″), the materials were rapidly dried by silica gel, and a voucher specimen (with collection numbers of YFL17102907) has been deposited in the Herbarium of Guizhou University of Traditional Chinese Medicine (GZYGH), Guiyang, China. The genomic DNA was extracted from leaves with the modified CTAB method (Doyle and Doyle Citation1987). Sequencing was performed on an Illumina HiSeq X-Ten to generate approximately 5‒70 million paired-end 150 bp reads at Beijing Genomics Institute (BGI, Wuhan, China). The GetOrganelle pipeline (Jin et al. Citation2018) was used to de novo assemble plastomes. In this pipeline, plastomic reads were extracted from total genomic reads and were subsequently assembled using SPAdes version 3.10 (Bankevich et al. Citation2012). Genes were annotated using PGA (Qu et al. Citation2019) and Geneious 11.0.3 (Kearse et al. Citation2012) with the published plastome of E. lishihchenii (GenBank accession number: NC_029944) as the reference. Transfer RNAs (tRNAs) were confirmed by tRNAscan-SE 2.0 (Lowe and Chan Citation2016).
The cp genome of E. dewuense (GenBank accession number: MN939629) is 157056 bp in length, displaying a quadripartite structure that contains a pair of inverted repeats (IR) regions (20820 bp), separated by a large single-copy (LSC) region (88346 bp) and a small single-copy (SSC) region (26768 bp). There are 130 genes reported, including 7 rRNA genes, 37 tRNA genes, and 86 protein-coding genes. The overall GC content of the cp genome was 38.8%, the GC content in LSC, SSC and IR regions was 37.4%, 36.6% and 43.1%, respectively. A total of 20 genes (trnK-UUU, trnI-GAU × 2, trnA-UGC × 2, trnG-UCC, trnV-UAC, trnL-UAA, rpoC1, ndhB × 2, ndhA, rpl16, rpl2, petB, atpF, petD, rps16, rps12 × 2) contained 1 intron, and 2 genes (clpP, ycf3) contained 2 introns.
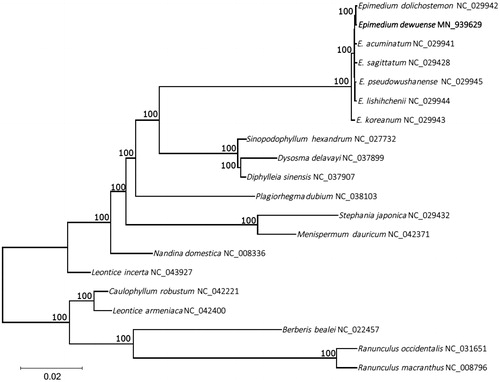
For phylogenetic analysis, a maximum-likelihood (ML) tree was constructed with 1000 bootstrap replicates using RAxML software (Stamatakis Citation2014). A subset of 16 species from the family Berberidaceae was included, with 4 species from Menispermaceae and Ranunculaceae as outgroup. The phylogenetic analyses showed that E. dewuense is placed under the the family Berberidaceae, clustered together with other Epimedium species, and E. dolichostemon were clustered together, indicating that the evolutionary relationship between them is closer (). The cp genome of E. dewuense provides valuable information and genomic resources regarding the taxonomy and phylogeny of Berberaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, ... & Pyshkin AV. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle J. J, Doyle J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bul. 19:11–15.
- He S. Z, Xu W. F. 2003. A new species of Epimedium (Berberidaceae) from Guizhou, China. Acta Botanica Yunnanica. 25:281–282.
- He S. Z. 2014. Color map of the genus Epimedium in China. Guiyang: Guizhou Science and Technology Press. p. 6.
- Jiang J, Zhao B. J, Song J, Jia X. B. 2016. Pharmacology and clinical application of plants in Epimedium L. Chin Herb Med. 8(1):12–23.
- Jin J. J, Yu W. B, Yang J. B, Song Y, Yi T. S, Li D. Z. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular cp genome using genome skimming data. BioRxiv:256479. doi:10.1101/256479
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe T. M, Chan P. P. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Ma H, He X, Yang Y, Li M, Hao D, Jia Z. 2011. The genus Epimedium: an ethnopharmacological and phytochemical review. J Ethnopharmacol. 134(3):519–541.
- Qu X. J, Moore M. J, Li D. Z, Yi T. S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu X, Li D X, Deng W L. 2010. Pharmacological progress of Epimedium on reproductive and endocrine system. Chin J Exp Trad Med Formulae. 16(8):223–227.
- Ying J, Boufford D E, Brach A R. 2000. Epimedium Linnaeus. Flora China. 29:262–298.
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee S-C, Yang T-J, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306.