Abstract
The Athetis pallidipennis belongs to Noctuidae in Lepidoptera. The complete mitogenome of A. pallidipennis was described in this study, which is typical circular duplex molecules and 15,344 bp in length, containing the standard metazoan set of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and an A + T-rich region. The gene order is same as other lepidopterans. Except for cox1 started with CGA, all other PCGs started with the standard ATN codons. Most of the PCGs terminated with the stop codon TAA, whereas cox2 and nad4 have the incomplete stop codon T––. The phylogenetic tree showed that A. pallidipennis and another species belong to the same Genus, which are clustered into a clade.
Athetis pallidipennis Sugi [1982] is mainly distributed in Japan, South Korea, and the Far East of Russia. New records of the species have been found in China in recent years (Zhang et al. Citation2017). We sequenced the complete mitogenome of A. pallidipennis in this study, which would facilitate the researches of the phylogeography and phylogenic relationship of Lepidoptera.
In this study, the samples were collected by light trapping in Taiyuan city of China (37°83′33″N, 112°66′61″E) in July 2019, some of these specimens were immediately frozen in −80 °C on board for mitogenome analysis, and others were preserved by spreading wings in the Shanxi Insect Herbarium, Shanxi Academy of Agricultural Sciences, and their numbers is 2019TYKD1520-1528. Total genomic DNA was extracted from tail tip using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). The mitogenome was sequenced using Illumina Hiseq 4000. Gene annotation was performed and circularity was checked using the MITOS2 webserver (Bernt et al. Citation2013, http://mitos.bioinf.uni-leipzig.de/).
The mitochondrial genome of A. pallidipennis has a total length of 15,344 bp (GenBank accession No. MT040606), consisting of 13 PCGs, 22 tRNA, two rRNA genes, and an A + T-rich region. The major strand encodes a larger number of genes (nine PCGs and 14 tRNAs) than the minor strand (four PCGs, eight tRNAs, and two rRNA genes). Gene content and arrangement are highly conserved and typical of Lepidoptera (Wu, Zhao, Su, He, et al. Citation2016; Wu, Zhao, Su, Luo, et al. Citation2016). The mitogenome is highly biased toward A/T, contains 41.47% T, 39.53% A, 11.31% C, and 7.69% G, which is a feature commonly present in insects (Boore Citation1999).
All of the protein-coding genes have ATN as the start codon except for cox1, which starts with CGA. Eleven PCGs have the common stop codon TAA. However, cox2 and nad4 have the incomplete stop codon T––. All tRNAs exhibit typical clover-leaf secondary structure, except for tRNA-Ser(AGN) lacking the DHU arm, which is common in Lepidoptera insects (Garey and Wolstenholme Citation1989). The 16S rRNA is 1315 bp in length, and the 12S rRNA is 766 bp in length. The A + T-rich region is 338 bp long located between 12S rRNA and tRNA-Met. There is a motif ATAGA in downstream of 12S rRNA followed by a 19 bp Poly-T stretch.
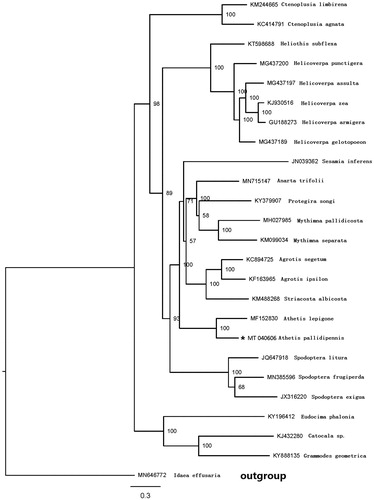
The phylogenetic position of A. pallidipennis was inferred using sequences of the 13 PCGs of 25 species. Twenty-four of them belong to Noctuidae, and Idaea effusaria from Geometridae (which is used as outgroup) (). The sequences were aligned with MAFFT v7.2 software (Katoh and Standley Citation2013). The evolutionary analyses were conducted with RAxML v8.2.10 (Stamatakis Citation2014) on the CIPRES Science Gateway (Miller et al. Citation2010). The result showed that A. pallidipennis and Athetis lepigone belong to the Genus Athetis, which are clustered into a clade.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of A. pallidipennis was deposited in GenBank under the accession number MT040606.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Garey JR, Wolstenholme DR. 1989. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 28(5):374–387.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14. New Orleans (LA): Institute of Electrical and Electronics Engineers (IEEE); p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu YP, Zhao JL, Su TJ, He QS, Xie JL, Zhu CD. 2016. The complete mitochondrial genome of Carposina sasakii (Lepidoptera: Carposinidae). Mitochondrial DNA. 27(2):1432–1434.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176.
- Zhang JH, Ding JY, Wu CS, Yao DD, Cui JC, Wang S, Duan YH, Gu PY. 2017. Three new record species of Noctuidae and one new record species of Crambidae from China (Lepidoptera). Plant Protection. 43(3):199–202.