Abstract
Lilium pumilum Redouté, belonging to Liliaceae, has important ornamental and medicinal values. In the present study, we sequenced the complete chloroplast genome of L. pumilum by Illumina Hiseq X Ten and PacBio RS technologies firstly. The genome size of L. pumilum was 152,591 bp, with typical quadripartite circle structure: one large single copy (LSC, 82,013 bp), one small single copy (SSC, 17,594 bp), and a pair of inverted repeat regions (IRs, 26,492 bp). The overall GC content was 37.02%. The genome contained 130 genes, including 84 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic analysis revealed that 23 Lilium species grouped into two branches, and L. pumilum was related to L. amabile and L. callosum. The present study could afford crucial genetic information for further researches on the genus and related genera.
The Lilium genus is comprised of 115 species that are mainly distributed throughout East Asia and North America (Liang and Tanmuram Citation2000). Many species of Lilium have high ornamental and medicinal values (Man and Samant Citation2011). Based on previous studies, no more than 30 Lilium species were recorded in GenBank. Thus, there was an urgent need for more genetic information about Lilium species. In the present study, we reported the complete chloroplast genome of Lilium pumilum, which could enrich the genetic information for further researches on the genus and related genera.
Lilium pumilum originally from Xiaocheng village, Qiankunwan town, Yan’an, China (N36°88′24.88″, E110°18′96.82″). Plantlets of L. pumilum (voucher number: SD_JBZ) were obtained from in vitro Shandandan germplasm preserved from the Conversation and Utilization of Regional Biological Resources, Yan’an, China.
Fresh clean leaves were selected and used for DNA extraction. Total genomic DNA was extracted using Plant Chloroplast DNA Column Extraction Kit (BNT120406, Baiao Laibo Technology Co., Ltd. Beijing, China). Then, genomic DNA was sequenced using PacBio RS and Illumina Hiseq X Ten platforms. SOAPdenovo v2.04 (Luo et al. Citation2012) was used for assembling after quality assessment. The sequencing data were subsequently aligned to PacBio RS sequencing data to remove a single base and compilation errors using blasR software (Sergey et al. Citation2012). The corrected data were assembled using Canu v1.5 (https://canu.es/) (Sergey et al. Citation2012). The whole chloroplast genome sequence of L. pumilum was deposited to NCBI (https://www.ncbi.nlm.nih.gov/genbank/) database with the accession number of MK954109.
Illumina Hiseq X Ten produced 5,537 Mb raw data, with an average Q20 > 90%. PacBio RS sequencing produced 9,912,133 bp subread bases, with an N50 of 9,693 bp. The chloroplast genome size of L. pumilum was 152,591 bp, with typical quadripartite structure: one large single copy (LSC, 82,013 bp), one small single copy (SSC, 17,594 bp), and a pair of inverted repeat regions (IRs, 26,492 bp). The total GC content of 37.01%. A total of 130 genes were detected, including 84 protein-coding genes, 38 tRNA genes, and eight rRNA genes. After removing duplicates, 78 protein-coding genes, 28 tRNA genes, and four rRNA genes remained.
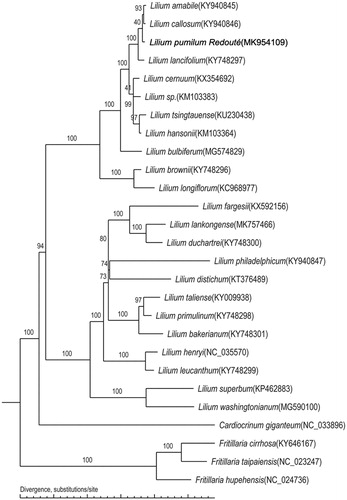
The phylogenetic tree of L. pumilum was constructed using the whole chloroplast genomes of 23 Lilium species and four outgroup species. The phylogenomic relationship was inferred using the Maximum-Likelihood (ML) method based on the general time-reversible (GTR) + G substitution model in PhyML 3.0 (Larkin et al. Citation2007; Guindon et al. Citation2010), with 1000 bootstrap replicates (Letunic et al. Citation2016). There were 13 nodes with support values of 100% and 20 nodes with support values greater than 90%. The phylogenetic tree showed that 23 Lilium species were grouped into two branches, and L. pumilum was related to L. amabile and L. callosum ().
Acknowledgments
Authors would to thank the company of Yan’an Shandan Biotechnology Co., Ltd. and Shanghai BIOZERON Biological Technology Co., Ltd for technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Letunic I, Bork P, Interactive Tree of Life (ITOL) v3. 2016. An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Liang SY, Tanmuram MN. 2000. Lilium. Flora of China. vol. 24. Beijing: Science Press; p. 135–149.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Man SR, Samant SS. 2011. Population biology of Lilium polyphyllum D. Don ex Royle-A critically endangered medicinal plant in a protected area of Northwestern Himalaya. J Nat Conserv. 19(3):137–142.
- Sergey K, Schatz MC, Walenz BP, Jeffrey M, Howard JT, Ganeshkumar G, Zhong W, Rasko DA, W Richard M, Jarvis ED. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 30(7):693–700.