Abstract
Axonopus compressus (Sw.) Beauv. is one of the most important warm-season turgrass, widely distributed in tropical regions of South China. In this study, we sequenced the complete chloroplast genome of A. compressus on the Illumina HiSeq Platform. The chloroplast genome is 139,227 bp in length, with a typical quadripartite structure and consisting of a pair of inverted repeat (IR) regions (22,753 bp) separated by a large single copy (LSC) region (81,111 bp) and a small single copy (SSC) region (12,610 bp). The overall GC content was 38.63%. A total of 138 genes were annotated, including 89 protein-coding genes, 41 tRNA genes, and eight rRNA genes. Phylogenetic analysis suggested Axonopus compressus (Sw.) Beauv. is sister to Axonopus ramosus plus Axonopus fissifolius.
Axonopus compressus (Sw.) Beauv. is a creeping, stoloniferous, perennial, warm-season grass that is adapted to humid tropical and subtropical climates (Zhang et al. Citation2020). Because A. compressus grows quickly and is tolerant to heat, shade, and drought stress, it is popular in public lawns, used to promote water and soil conservation, and planted as slope protection to prevent erosion along highways throughout southern China (Xi et al. Citation2006). However, its phylogenetic position is unclear in previous studies due to the lack of genomic resources of Axonopus genus. The Chloroplast genome is valuable in plant systematics research due to its highly conserved structures, uniparental inheritance, and haploid nature (Fu et al. Citation2016). Chloroplasts are an extremely important organelle to Earth creatures, and their genomes could be smartly engineered to confer useful traits (Jin and Daniell Citation2015). Consequently, the genetic and genomic information is urgently needed to promote its systematic research and utilization of A. compressus resources. In this study, we firstly report the complete chloroplast (cp) genome of A. compressus to provide useful genomic information for further research.
The plant material of A. compressus was collected from Hainan university Danzhou Campus, Haikou, Hainan province, China. The voucher specimen is kept at Hainan of University (specimen code HD20180920).The total genomic DNA was extracted from fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The original reading was filtered by CLC Genomics Workbench v9, and filtered sequences were assembled using the program SPAdes assembler v3.10.1 (Bankevich et al. Citation2012). Finally, CpGAVAS was used to annotate the gene structure (Liu et al. Citation2012). The obtained sequence was submitted to GenBank under the accession number MN989413.
The complete chloroplast genome of A. compressus was determined to comprise a 139,227 bp double stranded, circular DNA, which containing two inverted repeat (IR) regions of 22,753 bp, separated by large single-copy (LSC) and small single-copy (SSC) regions of 81,111 bp and 12,610 bp, respectively. The overall GC content of the chloroplast genome was 38.63%, while the corresponding values of the LSC, SSC, and IR regions were 36.53%, 32.83%, and 43.98%, respectively. A total of 138 functional genes were annotated, including 89 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Ten protein-coding genes, eight tRNA genes and four rRNA genes were duplicated in IR regions. Fourteen genes (eight protein-coding genes and six tRNA genes) contained one intron and two genes (rps12 and ycf3) contained two introns.
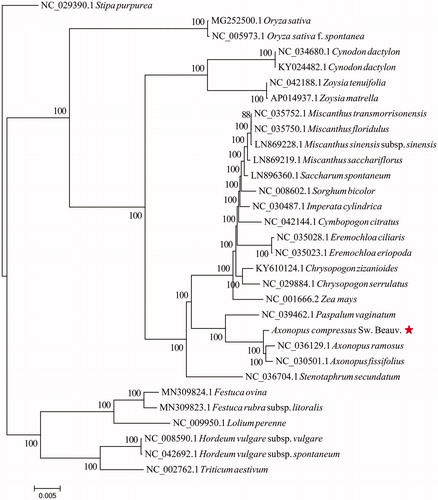
The maximum likelihood phylogenetic tree was generated based on the plastid genome of A. compressus and other 30 species of the Poaceae. Alignment was conducted using MAFFT v7.307 (Katoh and Standley, Citation2013). The phylogenetic tree was built using RAxML (Stamatakis Citation2014) with bootstrap set to 1000. The maximum likelihood (ML) phylogenetic tree suggested that Axonopus compressus (Sw.) Beauv. is sister to Axonopus ramosus plus Axonopus fissifolius with strong support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Fu PC, Zhang YZ, Geng HM, Chen SL. 2016. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 4:e2540.
- Jin S, Daniell H. 2015. The engineered chloroplast genome just got smarter. Trends Plant Sci. 20(10):622–640.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30(4):772–780.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xi JB, Chen P, Zheng YZ, Yang ZY. 2006. An investigation of Axonopus compressus germplasm resources in China. Acta Pratac Sin. 15:93–99.
- Zhang W, Ulrike D, Pedro WC, Johannes ZG, Niu XL, Lin JM, Li YT. 2020. Anthracnose Disease of Carpetgrass (Axonopus Compressus) Caused by Colletotrichum ainanense Sp. Nov. Plant Disease. DOI:10.1094/PDIS-10-19-2183-RE