Abstract
The complete mitochondrial genome (mitogenome) of the Bactrocera rubigina (Diptera: Tephritidae: Dacinae) was sequenced and annotated. The mitochondrial genome was 15,285 bp (GenBank No. MN714223), containing 73.1% of A + T with A 39.3%; C 16.9%; G10.1% and T 33.8%, respectively, which is the classical structure for insect mitogenome. All PCGs started with ATN and stopped with TAD. The phylogenetic tree in the current study confirms that B. rubigina clustered with other Bactrocera species, and this study would enrich the mitogenomes of the fruit flies.
The genus Bactrocera (Dipeter, Tephritidae, Dacinae) was found by Macquart in 1983 for a single species of Bactrocera longiwnis. Presently, more than 470 species of the genus are known from different regions of Asia, Australia, and South Pacific, with a few species distributed in Southern Africa and Hawaiian Islands (Wang Citation2008; You Citation2014). Some species of the genus are important pests in fruit and vegetable crops. In current study, the complete mitochondrial genome (mitogenome) was sequenced and determined using next-generation sequencing method for the first time, which obtained for fruit flies will facilitate future studies on species identification, population genetics, and phylogenetic analysis of the subfamily Dacinae.
The genome DNA was extracted from male adult of B. rubigina which was collected from Shiban town, Guizhou province, China (E 106°41′, N 26°26′), in June 2019 and voucher specimen’s genome DNA are deposited in the Research Center for Guizhou Characteristic Fruits and its Products of Mountainous Regions, Guizhou Light Industry Technical College, label number is GCZX-137. These sequences were assembled using Geneious Primer (Kearse et al. Citation2012), version 10.2.3 (http://www.geneious.com/). In addition, all tRNAs were found by MITOS server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and tRNA scan-SE server (Lowe and Chan Citation2016) for annotation. The neighbor-joining (NJ) tree was constructed to investigate the molecular taxonomic position of B. rubigina based on nucleotide sequences of 13 protein-coding genes and 2 rRNA genes using MEGA 6.0 (Tamura et al. Citation2013) from alignments created by the MAFFT (Katoh and Standley Citation2013).
The complete mitogenome of B. rubigina was 15,285 bp (GenBank No. MN714223), containing 22 transfer RNA genes (tRNAs, 1468 bp), 13 protein-coding genes (PCGs, 11,336 bp), 2 ribosomal RNA genes (rRNAs, 2115 bp), and non-coding regions (control regions, 282 bp). The composition of the whole genome contained 39.3% A, 16.9% C, 10.1% G, and 33.8% T, showing an obvious A + T bias (73.1%). The AT-skew (0.075) for the whole mitogenome is slightly positive, but negative for GC-skew (−0.252). All PCGs started with ATN (ATA/ATG/ATT/ATC) and stopped with TAD (TAA/ATG/TAT).
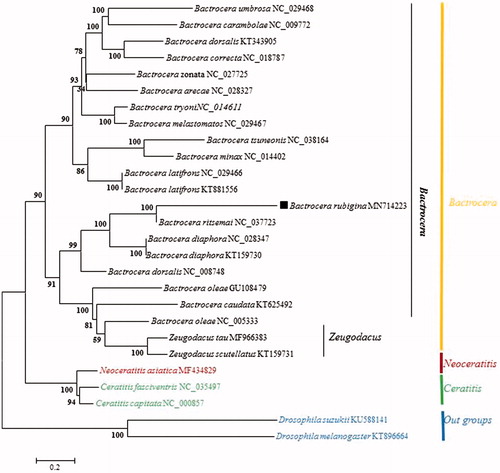
The phylogenetic relationships of B. rubigina were reconstructed using the neighbor-joining (NJ) method with 1000 bootstrap replicates based on concatenated nucleotides of the 13 PCGs and 2 rRNAs with 12,804 bp, Dosophila suzukii and Drosophila melanogaster were used as outgroup (). The phylogenetic tree confirmed that all the species of the genus Bactrocera formed a clade, B. rubigina was closely related to B. ritsemai and B. Diaphora with strongly supported (bootstrap support value = 100). Presentluy, the studies recorded for Bactrocera was limited and we hope that our data can be useful for further study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe TM, Chan PP. 2016. Trna scan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang XJ, Xiao S, Chen XL, Long R, Zhang CL. 2008. Two newspecies of the genus Bactrocera Macquart (Diptera, Tephritidae) from Yunnan, China. Acta Zootaxonomica Sinica. 33 (1):73–76.
- You H, Zhou LB, Deng YL, Chen GH. 2014. Identification of Bactrocera species from the border areas of Yunnan province, southwestern China using DNA barcoding (Diptera: Tephritidae). Acta Entomologica Sinica. 57(11):1343–1350.