Abstract
The complete mitochondrial genome of Penicillium sp. D1806 isolated from Oryza sativa seeds is reported on the basis of the Illumina sequencing data. Its circular mitogenome is 27,461 bp in length, containing 15 protein-coding genes (PCGs), 1 ORFs, 2 ribosomal RNA (rns and rnl) genes, and 24 transfer RNA (tRNA) genes. The overall base composition is as follows: 36.2% A, 37.1% T(U), 11.8% C, 14.9% G, with a low GC content of 26.7%. Phylogenetic analysis shows that Penicillium sp. D1806 is clustered in the genus Penicillium of Aspergillaceae and forms a separate clade with strong statistical support. This study contributes to our understanding of systematics and evolutionary biology of the filamentous fungi in Aspergillaceae (Eurotiomycetes, Ascomycota).
Penicillium has a worldwide distribution and is one of the most common fungi occurring in a diverse range of habitats, such as soil, vegetation, air, indoor environments, and food products (Visagie et al. Citation2014). It has a large economic impact on human life. Many species cause devastating rots as pre- and postharvest pathogens on food crops, as well as producing divers mycotoxins (Frisvad et al. Citation2004; Frisvad and Samson Citation2004; Pitt and Hocking Citation2009; Samson et al. Citation2010). Members of P. section Citrina are very abundant (nearly 40 species) and are well known for their ability to produce the mycotoxins citrinin and citreoviridin (Houbraken et al. Citation2011). They are very common in soil, but are also usually isolated from indoor environments and foodstuffs (Pitt and Hocking Citation2009; Samson et al. Citation2010). In this study, we determined the complete mitogenomic sequence of Penicillium sp. D1806 belonging to the section Citrina which might contribute to the understanding of systematics and evolutionary biology in Penicillium.
Penicillium sp. D1806 was isolated from Oryza sativa seeds collected from Panlong district of Kunming in Yunnan province, southwestern China (25°86′N, 102°65′E, alt. 2050 m). Morphological observation and ITS sequences analysis showed that the strain belonged to P. section Citrina. The strain D1806 was deposited at the Institute of Biotechnology and Germplasm Resources, Yunnan Academy of Agricultural Sciences, China. Mycelia cultured on PDA at 25 °C for 14 days were prepared to extract total genomic DNA using DNeasy Plant Genomic DNA Purification Mini Kit (QIAGEN). The whole-genome sequencing was performed using the Illumina sequencing platform (HiSeq-PE150). The software SPAdes v. 3.11.0 was used to assemble mitogenome of Penicillium sp. D1806 (Bankevich et al. Citation2012). MFannot tool and ARWEN web server were used to annotate the mitogenome. The Organellar Genome DRAW tool was used to draw the mitogenomic circular map (Lohse et al. Citation2007).
The circular mitogenome of Penicillium sp. D1806 is 27,461 bp in length, containing 15 protein-coding genes (PCGs), 1 ORFs (ORF349), 2 ribosomal RNA (rns and rnl) genes, and 24 transfer RNA (tRNA) genes. There are three trnM-CAT genes in the set of 24 tRNAs. The total length of the 15 PCGs (atp6, atp8–9, cob, rps3, cox1–3, nad1–6, and nad4L) and ORF349 (a hypothetical protein) is 15,158 bp, which accounts for 55.20% of the whole mitogenome. The lengths of 24 transfer RNA (tRNA) genes are ranging from 71 to 86 bp, among which the shortest is trnG-ACC gene (71 bp) and the longest is trnS-TGA gene (86 bp). The sizes of rns and rnl are 1522 bp and 4724 bp, respectively. The overall base composition of Penicillium sp. D1806 is as follows: 36.2% A, 37.1% T(U), 11.8% C, 14.9% G, with a low GC content of 26.7%. The complete mitogenome sequence of Penicillium sp. D1806 was submitted to GenBank database under accession number no. MN960691.
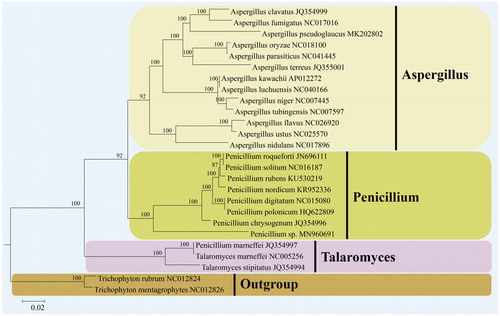
Fourteen concatenated mitochondrial PCGs were employed to carry out phylogenetic analysis to determine the systematic position of Penicillium sp. D1806. Sequence alignment and phylogenetic analysis were conducted as described by Wang et al. (Citation2018). Penicillium sp. D1806 was clustered in the genus Penicillium (Aspergillaceae, Eurotiomycetes) and formed a separate clade with high credible support by Bayesian inference posterior probabilities (BI-PP = 100%), having the distant phylogenetic relationship with other species of Penicillium ().
Figure 1. Phylogenetic position of Penicillium sp. D1806 in the family Aspergillaceae based on the Bayesian inference analysis of 14 concatenated mitochondrial protein-coding genes (PCGs). The 14 PCGs include subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6). Bayesian inference posterior probabilities are shown above the internodes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Frisvad JC, Samson RA. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol. 49:1–174.
- Frisvad JC, Smedsgaard J, Larsen TO, Samson RA. 2004. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 49:201–241.
- Houbraken J, Frisvad JC, Samson RA. 2011. Taxonomy of Penicillium section Citrina. Stud Mycol. 70:53–138.
- Lohse M, Drechsel O, Bock R. 2007. Organellargenomedraw (ogdraw): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Pitt JI, Hocking AD. 2009. Fungi and food spoilage. New York: Springer.
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. 2010. Food and indoor fungi. Utrecht: CBS KNAW Biodiversity Center.
- Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol. 78:343–371.
- Wang YB, Nguyen TT, Dai YD, Yu H, Zeng WB, Wu CK. 2018. Molecular phylogeny and morphology of Ophiocordyceps unituberculata sp. nov. (Ophiocordycipitaceae), a pathogen of caterpillars (Noctuidae, Lepidoptera) from Yunnan, China. Mycol Prog. 17(6):745–753.
