Abstract
We sequenced the complete mitochondrial genome of Hamamelistes spinosus using the shotgun genome-skimming method. The mitogenome of H. spinosus is 15,089 bp in length with a much higher A + T content of 81.8% than that of G + C (18.2%) and consists of 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes and 1 control region. All the 13 PCGs initiate with a typical ATN and end with TAA except for two genes COX1 and ND4, which terminate with a single T. All the tRNAs form a classical clover-leaf secondary structure except for tRNA-Ser (AGN), which does not have the dihydrouridine (DHU) arm. In addition, we constructed the phylogenetic tree of Aphididae with two species Adelges laricis and Daktulosphaira vitifoliae as outgroups and the results showed that Hamamelistes spinosus is sister to Hormaphis betulae and they are closely allied with Neothoracaphis yanonis.
Hamamelistes spinosus belongs to the tribe Hormaphidini (Hemiptera: Aphididae: Hormaphidinae), and has alternate hosts with the American witch-hazel (Hamamelis virginiana of Hamamelidaceae) as the primary host and the river birch (Betula nigra of Betulaceae) as the secondary host to finish its life cycle (Remaudière and Remaudière Citation1997). The aphid species feeds on the leaves of the primary host to induce galls, which is conducive to aphids for the efficient interception of nutrients and self- protection (Inbar et al. Citation2004). Hormaphidinae contains only three tribes and 49 genera (Favret Citation2020), however, so far, only one complete mitochondrial genome (mitogenome) of Hormaphis betulae was reported in this subfamily (Li et al. Citation2015). Herein, we sequenced the complete mitogenome of Hamamelistes spinosus, and constructed the phylogenetic analyses with other Aphididae species by combining the data from Genbank.
We collected the sample of Hamamelistes spinosus from a gall on Hamamelis virginiana in Polk County (34°27′55″N, 94°31′35″W), AK, USA, in 2009. All the aphid individuals from a gall are parthenogenetic, and we deposited some individuals from the same gall as the voucher specimen at the animal herbarium of School of Life Science, Shanxi University, China (voucher no Ren_A405). We sequenced the mitogenome of H. spinosus by the shotgun genome-skimming method on an Illumina HiSeq 4000 platform (Zimmer and Wen Citation2015). The mitogenomic sequence of H. spinosus was assembled by SPAdes v. 3.7.1 (Bankevich et al. Citation2012) and annotated within Geneious v. 11.0.3 using the complete mitogenome of Hormaphis betulae (GenBank Accession no. KT875793) as the reference.
The complete mitogenome of Hamamelistes spinosus is a circular double-stranded DNA with 15,089bp in length (GenBank accession no. MT010853), which consists of 13 protein-coding genes (PCGs, COX1-COX3, ND1-ND6, ND4L, ATP6, ATP8, Cyt b), two rRNAs (12S and 16S rRNA), 22 tRNAs and one control region (D-loop). The content of A + T (81.8%) is significantly higher than that of G + C (18.2%). Most of genes encoded on H-strand except for the 13 genes (ND1, ND4L, ND5, 12S rRNA, 16S rRNA, tRNA-Cys, tRNA-Phe, tRNA-Gln, tRNA-Tyr, tRNA-Pro, tRNA-Leu, tRNA-Val) encoded on L-strand. All the 13 PCGs are initiated with ATN, among which six genes (ATP6, ATP8, ND1, ND4-ND6) started with ATT, whereas, five genes (COX1, COX2, ND2, ND3, ND4L) initiated with ATA, and only two genes (COX3 and Cyt b) started with ATG. Eleven PCGs terminate with TAA, whereas COX1 and ND4 terminate with a single T. We predicted the secondary structure of tRNA by MITOS v.2 WebServe (Bernt et al. Citation2013) and found that all the tRNAs form a classical clover-leaf secondary structure except for tRNA-Ser (AGN), which loses a dihydrouridine (DHU) arm.
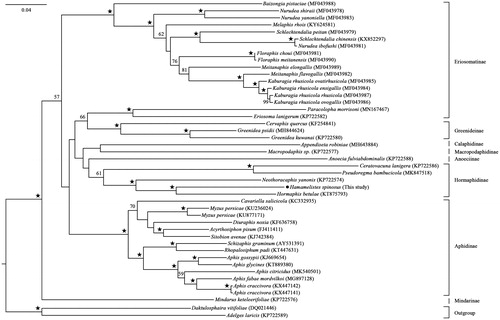
We downloaded the mitogenomic sequences of Aphididae species from GenBank with two species Adelges laricis and Daktulosphaira vitifoliae as outgroups to construct the maximum likelihood phylogenetic relationship of the Aphididae including H. spinosus by RAxML v. v.8.2.10 (Stamatakis Citation2014). The tree supported the monophyly of the subfamily Hormaphidinae, and Hamamelistes spinosus is sister to Hormaphis betulae, which is closely allied with the species Neothoracaphis yanonis ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 69(2):313–319.
- Favret C. 2020. Aphid species file. Version 5.0/5.0. http://Aphid.SpeciesFile.org. [retrieval 2020 Feb 6].
- Inbar M, Wink M, Wool D. 2004. The evolution of host plant manipulation by insects: molecular and ecological evidence from gall-forming aphids on Pistacia. Mol Phylogenet E. 32(2):504–511.
- Li Y-Q, Chen J, Qiao G-X. 2015. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae). Mitochondr DNA A DNA Mapp Seq Anal. 28 (2):265–266.
- Remaudière G, Remaudière M. 1997. Catalogue of the world’s Aphididae, Homoptera, Aphidoidea. Paris, France: Institut National de la Recherche Agronomique.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zimmer EA, Wen J. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. Jnl of Sytematics Evolution. 53(5):371–379.