Abstract
Mitochondrial sequences are commonly utilized as genetic makers in molecular genetic studies for Holothuroidea species. In this study, the complete mitochondrial genome (mitogenome) of Colochirus robustus is reported for the first time. These data demonstrate that the C. robustus mitochondrial was a 17,171 bp circular molecule and encodes the typical 37 metazoan mitochondrial genes [13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs)] genes, and a putative control region. The genome composition was highly A + T biased 67.55% and showed positive AT-skew (0.182) and negative GC-skew (−0.314). The complete mitogenome provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for holothurians.
Holothuroids, also known as sea cucumbers, are an abundant and diverse group, which includes more than 1400 species occurring from the intertidal to the deepest oceanic trenches (Purcell et al. Citation2012; Gallo et al. Citation2015; Mu et al. Citation2018). Identification of sea cucumbers was usually done using morphological characteristics, which is a very time-consuming method that requires highly skilled experts (Kamarudin & Rehan Citation2015; Madduppa et al. Citation2017). Many of their morphological characters show a high degree of variation some of which may arise as a result of artifacts of methods used for collection and preservation of specimens (Solís-Marín et al. Citation2004). So multiple incongruences have been found between morphology based taxonomic systems and molecular systematic reports (Littlewood et al. Citation1997; Miller et al. Citation2017). However, the complete mitochondrial sequencing shows a great potential to both resolve disputed taxonomic issues and to infer phylogenetic relationships among holothurians (Wang et al. Citation2018; Utzeri et al. Citation2018; Yang et al. Citation2019).
In this study, muscle sample of a C. robustus individual was collected from Dongshan, Fujian Province of China (23°43′23″N, 117°29′19″E). The specimen was preserved in the Culture Collection of Sea cucumber at the Fisheries research institute of Fujian of China (specimen number: DS-191103) and stored at −80 °C for DNA isolation. Total DNA was extracted from the muscle tissues and using the Aidlab Genomic DNA Extraction Kit (Aidlab Biotech, Beijing, China). The mitogenomes of C. robustus were sequenced by next-generation sequencing (Illumina HisSeq 4000; Shanghai Origingene Biopharm Technology Co. Ltd., Shanghai, China). Clean data without sequencing adapters were de novo assembled using the NOVO Plasty software (Dierckxsens et al. Citation2017). The complete mitogenome was annotated using MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) with the selection of invertebrate mitochondrial genetic code. Transfer RNA (tRNA) genes were identified using tRNAscan-SE online (Lowe and Chan Citation2016). A phylogenetic analysis was performed with nine complete mitogenomes of holothuroids, including C. robustus, and two Ophiuroidea, Amphipholis squamata (FN562578.1) and Amphiura digitula (MH791160.1) were used to outgroups for this analysis.
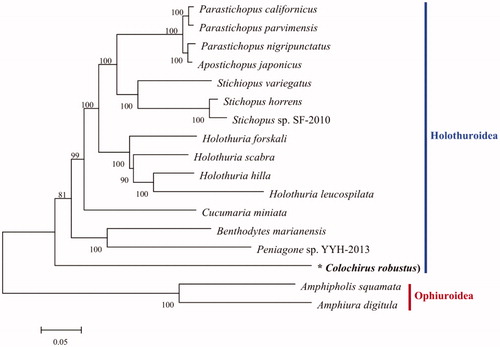
The complete mitogenome of C. robustus (GenBank accession No. MN966676) was 17,171 bp in length and contained 13 PCGs, 22 tRNA genes, and 2 rRNA genes. The gene order and direction of PCGs and rRNAs were identical to those of the other holothuroids complete mitogenomes (Lee & Shin Citation2019; Yang et al. Citation2019). Overall, nucleotide base composition of C. robustus mitogenome was A 39.94%, T 27.62%, C 21.31% and G 11.13% with an overall A + T content of 67.56%, which is similar, but higher than Actinopyga echinites (62.9%) (Zhong et al. Citation2020). In the phylogenetic tree, C. robustus was grouped distinctly with the other species, forming an independent branches (). As the result, the mitochondrial genome sequence of C. robustus was the first sequenced mitogenome in Colochirus, which will be a resource for further phylogenetic studies of mitogenome in holothuroids.
Figure 1. Phylogeny of holothurians based on amino acid sequences of 13 mitogenome PCGs using ML methods. Numbers on branches indicate bootstrap probability. The asterisks before species names indicate newly determined mitochondrial genome in this paper. The gene’s accession number for tree construction is listed as follows: Parastichopus californicus (KP398509), P. parvimensis (KU168761), P. nigripunctatus (AB525762), Apostichopus japonicus (FJ986223), Stichiopus variegatus (MN128376), S. horrens (HQ000092), Stichopus sp. SF-2010 (HM853683), Holothuria forskali (FN562582), H. scabra (KP257577), H. hilla (MN163001), H. leucospilata (MK940237), Cucumaria miniata (AY182376), Colochirus robustus (MN966676), Peniagone sp. YYH-2013 (KF915304), Benthodytes marianensis (MH208310), Amphipholis squamata (FN562578), Amphiura digitula (MH791160).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Gallo ND, Cameron J, Hardy K, Fryer P, Bartlett DH, Levin LA. 2015. Submersible- and lander-observed community patterns in the Mariana and New Britain trenches: Influence of productivity and depth on epibenthic and scavenging communities. Deep Sea Res Part I Oceanogr Res Pap. 99:119–133.
- Kamarudin KR, Rehan MM. 2015. Morphological and molecular identification of Holothuria (Merthensiothuria) leucospilota and Stichopus horrens from Pangkor Island, Malaysia. Trop Life Sci Res. 26(1):87–99.
- Lee T, Shin S. 2019. Complete mitochondrial genome of sea cucumber, Holothuria (Stauropora) pervicax (Holothuroidea, Holothuriida, Holothuriidae), from Jeju Island, Korea. Mitochondr DNA B. 4(1):1047–1048.
- Littlewood DTJ, Smith AB, Clough KA, Emson RH. 1997. The interrelationships of the echinoderm classes: morphological and molecular evidence. Biol J Linn Soc. 61(3):409–438.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Madduppa H, Taurusman AA, Subhan B, Anggraini N, Fadillah R, Tarman K. 2017. Short communication: DNA barcoding reveals vulnerable and not evaluated species of sea cucumbers (Holothuroidea and Stichopodidae) from kepulauan Serbu reefs, Indonesia. Biodiversitas. 18(3):893–898.
- Miller AK, Kerr AM, Paulay G, Reich M, Wilson NG, Carvajal JI, Rouse GW. 2017. Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol Phylogenet E. 111:110–131.
- Mu WS, Liu J, Zhang HB. 2018. Complete mitochondrial genome of Benthodytes marianensis (Holothuroidea: Elasipodida: Psychropotidae): Insight into deep sea adaptation in the sea cucumber. PLOS One. 13(11):e0208051.
- Purcell SW, Samyn Y, Conand C. 2012. Commercially important sea cucumbers of the world. Italy: FAO. Report No. 6. FAO species catalogue for fishery purposes; p. 102.
- Solís-Marín FA, Billett DSM, Preston J, Rogers AD. 2004. Mitochondrial DNA sequence evidence supporting the recognition of a new North Atlantic Pseudostichopus species (Echinodermata: Holothuroidea). J Mar Biol Ass U Ass. 84(5):1077–1084.
- Utzeri VJ, Ribani A, Bovo S, Taurisano V, Calassanzio M, Baldo D, Fontanesi L. 2018. Microscopic ossicle analyses and the complete mitochondrial genome sequence of Holothuria (Roweothuria) polii (Echinodermata; Holothuroidea) provide new information to support the phylogenetic positioning of this sea cucumber species. Mar Genomics. doi:10.1016/j.margen.2019.100735
- Wang Z, Wang Z, Shi X, Wu Q, Tao Y, Guo H, Ji C, Bai Y. 2018. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int Journal of Biol Macromol. 118(Pt A):31–40.
- Yang QH, Lin Q, Wu JS, Yang FY, Ge H, Qiu DG, Li ZQ, Lu Z, Li SK, Zhou C. 2019. The complete mitochondrial genome sequence of Stichopus variegatus (Echinodermata: Holothuroidea) and phylogenetic studies of Echinodermata. Mitochondr DNA B. 4(2):3244–3245.
- Yang QH, Lin Q, Yang FY, Wu JS, Lu Z, Li SK, Zhou C. 2019. Characterization of the complete mitochondrial genome of a holothurians species: Holothuria hilla (Holothuroidea: Holothuriidae). Mitochondr DNA B. 4(2):2847–2848.
- Zhong SP, HUang LH, Liu YH, Huang GP. 2020. The first complete mitochondrial genome of Actinopyga from Actinopyga echinites (Aspidochirotida: Holothuriidae). Mitochondr DNA B. 5(1):854–855.
