Abstract
The species Deutzia pilosa is well known as a commonly cultivated shrub and traditional Chinese herbal medicine. Here, we report the complete chloroplast genome of D. pilosa. It shows a total length of 156,983 bp with typical quadripartite structure, including the large single copy region (LSC) of 86,598 bp, the small single copy region (SSC) of 18,725 bp, and two separated inverted regions (IRs) of 25,830 bp, respectively. It contains 134 genes, including 89 protein-coding genes, 37 rRNA genes, and 8 tRNA genes. The overall GC content of the cp genome is 43.0%.
The genus Deutzia, with more than 60 species, belongs to the Trib. Philadelpheae of Hydrangeaceae and shows a distribution in East Asia, Mexico and Central America (Shumei Citation1993). Deutzia pilosa Rehd. is often cultivated as ornamental shrub in South China for their spotless, flawless and fascinating flowers. In the current study, we report the first complete chloroplast (cp) genome of D. pilosa, on the purpose of further phylogenetic reconstruction or biogeographic study of the Hydrangeaceae family.
One individual was collected from Emei Mts., Sichuan Province (Voucher No. FQ190401). Tender leaves were sampled and instantly put into the silica gel for drying and preservation. Total genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987). The paired-end (2 × 150 bp) library was sequenced by Illumina PE150 at Novogene Co. Ltd (Beijing, China). A total of 1.51 Gb clean reads were obtained after removing low-quality reads and adaptor sequences. The complete cp genome of D. pilosa was assembled using GetOrganelle and SPAdes (Bankevich et al. Citation2012), with manual adjustment and annotation using Geneious v8.0.2. The tRNA genes were annotated on ARAGORN (Laslett and Canback Citation2004). The cp genomes of ten species of Hydrangeaceae were downloaded from GenBank database, including Cardiandra moellendorffii (MN380653), Decumaria barbara (MN380684), D. sinensis (MN380685), Deinanthe caerulea (MN380658), Deutzia compacta (MN380704), Dichroa febrifuga (MN380702), Hydrangea aspera (MG524992), H. luteovenosa (MF370556), Philadelphus calvescens (MN380700) and Schizophragma hydrangeoides (KY412467). Paeonia lactiflora (MG897127) of Paeoniaceae was used as an outgroup. Totally, twelve cp genomes were aligned with online software MAFFT on CIPRES (https://www.phylo.org., Miller et al. Citation2015). Maximum likelihood (ML) analyses were performed using RAxML-HPC v.8 on XSEDE (https://www.phylo.org) with 1000 bootstrap replicates and the substitution GTR + I + G model (Stamatakis et al. Citation2008), the latter of which was determined by the Akaike information criterion (AIC) in jModeltest v2.1.10 (Darriba et al. Citation2012).
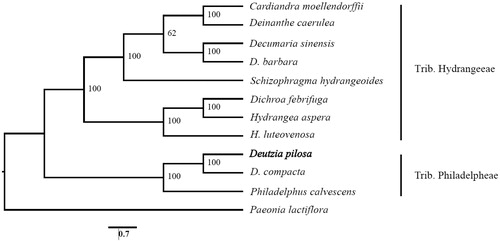
The D. pilosa cp genome has been deposited in GenBank (Accession No.: MN954672). The total length is 156,983 bp with the typical quadripartite structure. It consists of a pair of inverted regions (IRs) of 25,830 bp separated by large single-copy region (LSC) of 86,598 bp and small single copy region (SSC) of 18,725 bp, respectively. The overall GC content of the cp genome is 43.0%. The whole cp genome of D. pilosa contains 134 genes with 89 protein-coding genes (PCGs), 37 tRNA genes, and eight rRNA genes. Among these genes, 63 PCGs and 22 tRNA genes are located in the LSC region, while 12 PCGs and one tRNA genes occur in the SSC region. All these eight rRNA genes are duplicated in the IR regions. IR regions contain seven PCGs and seven tRNA genes, if counting only once. Among the annotated genes, eleven genes (atpF, ndhA, petB, petD, rpl16, rpoC1, rps16, trnaK-UUU, trnG-UCC, trnL-UAA, and trnV-UAC) contain one intron, while seven genes (clpP, ndhB, rpl2, rps12, trnA-UGC, trnL-GAU, and ycf13) possess two introns. The species D. pilosa together with D. compacta form a monophyly that is closely related with Philadelphus calvescens in the Trib. Philadelpheae ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bullet. 19:11–15.
- Laslett D, Canback B. 2004. ARAGORN, a program to the detection of transfer RNA and transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O'Leary MA. 2015. A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evol Bioinform Online. 11:EBO.S21501–48.
- Shumei H. 1993. The classification and distribution of genus Deutzia in China. J Chin Acad Sci. 31(2):105–126.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. System Biol. 57(5):758–771.