Abstract
The complete mitochondrial genome of the important black aspergilli species Aspergillus japonicus was determined using the next-generation sequencing technology. The circular mitogenome of A. japonicus YFCC 8801-W is 48,001 bp in length with a GC content of 27.1%, containing 14 protein-coding genes (PCGs), 4 ORFs, 24 transfer RNA (tRNA) genes and 2 ribosomal RNA (rns and rnl) genes. Phylogenetic analysis based on 14 PCGs showed that A. japonicus was clustered in the A. section Nigri clade of the genus Aspergillus (Aspergillaceae) and formed a separate clade with high credible support. This study provides valuable information for further exploration of mitochondrial evolution in the order Aspergillales.
Aspergillus is one of the most abundant and widely distributed genera and has a significant impact with applications in many areas. Among this lineage, the black aspergilli (A. section Nigri) are an important group in food mycology, medical mycology, and biotechnology. Aspergillus japonicus, belonging to the section Nigri that produces black colonies, has usually been isolated from soils, decomposing plant roots and leaves, predominantly in tropical and subtropical regions (Pasin et al. Citation2017). It was earlier reported to produce Asperparaline A, asperparaline B and C and showed paralytic activity against silkworms (Hayashi et al. Citation1997, Citation2000). Previous studies also showed that A. japonicus is a good producer of different enzymes, such as pectinases, β-fructofuranosidase, β-glucosidase, cellulases, tannases, phytases, lipases, xylanases and beta-xylosidases (Semenova et al. Citation2003, Citation2009; Maller et al. Citation2010; Facchini et al. Citation2011; Pasin et al. Citation2017). However, little is known about its genomic information. This study aims to report the complete mitogenomic characterization of A. japonicus and verify its phylogenetic position in the family Aspergillaceae.
Aspergillus japonicus strain YFCC 8801-W used in this study was isolated from house dust of The Third Affiliated Hospital of Kunming Medical University, Xishan district of Kunming in China (25°02′N, 102°40′E, alt. 1880 m). This strain was deposited at the Yunnan Fungal Culture Collection (YFCC), Yunnan University. Axenic cultures on PDA at 25 °C for 14 days were used to extract genomic DNA using DNeasy Plant Genomic DNA purification Mini Kit (QIAGEN). The whole-genome sequencing was performed on the Illumina sequencing platform (HiSeq-PE150). The sequencing data were assembled by the software SPAdes v. 3.11.0 (Bankevich et al. Citation2012). The mitogenome of A. japonicus YFCC 8801-W was annotated using MFannot tool and ARWEN web server, together with artificial correction technology. The mitogenomic circular map was drawn by the Organellar Genome DRAW tool (Lohse et al. Citation2007).
The complete mitogenomic sequence of A. japonicus YFCC 8801-W was submitted to GenBank database under accession number no. MN960692. The circular mitogenome is 48,001 bp in length with a GC content of 27.1%, containing 14 protein-coding genes (PCGs), 4 ORFs, 24 transfer RNA (tRNA) genes and 2 ribosomal RNA (rns and rnl) genes. The total length of 14 PCGs (atp6, atp8–9, cob, cox1–3, nad1–6 and nad4L) and 4 ORFs (ORF1010, ORF331, ORF158 and ORF597) is 31,866 bp, accounting for 66.39% of the whole mitogenome. The lengths of 24 transfer RNA (tRNA) genes are ranging from 71 to 85 bp. The lengths of rrns and rrnl are 1455 bp and 5271 bp, respectively.
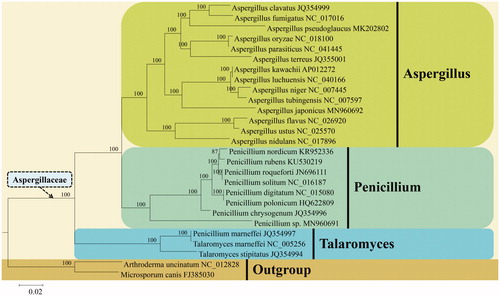
To verify the phylogenetic position of A. japonicus, 14 concatenated PCGs from 27 mitogenomes downloaded from NCBI were used for Bayesian inference phylogenetic analysis. Sequence alignment and phylogenetic analysis were conducted as described by Wang et al. (Citation2018). Phylogenetic tree showed that A. japonicus was clustered in the A. section Nigri clade within the genus Aspergillus of Aspergillaceae (Aspergillales, Eurotiomycetes) and formed a separate clade with high credible support by Bayesian inference posterior probabilities (PP = 100%) ().
Figure 1. Phylogenetic relationship between Aspergillus japonicus and its allies in the family Aspergillaceae based on the Bayesian inference analysis of 14 concatenated mitochondrial protein-coding genes (PCGs). The 14 PCGs are comprised of five subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, atp9), and NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6). Bayesian inference posterior probabilities are shown above the internodes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Facchini FDA, Vici AC, Benassi VM, Freitas LAP, Reis RA, Jorge JA, Terenzi HF, Polizeli M. 2011. Optimization of fibrolytic enzyme production by Aspergillus japonicus C03 with potential application in ruminant feed and their effects on tropical forages hydrolysis. Bioprocess Biosyst Eng. 34(8):1027–1038.
- Hayashi H, Nishimoto Y, Nozaki H. 1997. Asperparaline A, a new paralytic alkaloid from Aspergillus japonicus JV-23. Tetrahedron Lett. 38(32):5655–5658.
- Hayashi H, Nishimoto Y, Akiyama K, Nozaki H. 2000. New paralytic alkaloids, asperparalines A, B and C, from Aspergillus japonicus JV-23. Biosci Biotech Bioch. 64(1):111–115.
- Lohse M, Drechsel O, Bock R. 2007. Organellargenomedraw (ogdraw): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Maller A, Damasio ARL, Silva TM, Jorge JA, Terenzi HF, Polizeli M. 2010. Potential application in animal feed of phytase produced from agro-industrial residues by Aspergillus japonicus. J Biotechnol. 150:514–514.
- Pasin TM, Benassi VM, Heinen PR, de Lima Damasio AR, Cereia M, Jorge JA, de Moraes M. 2017. Purification and functional properties of a novel glucoamylase activated by manganese and lead produced by Aspergillus japonicus. Int J Bio Macromol. 102:779–788.
- Semenova MV, Drachevskaya MI, Sinitsyna OA, Gusakov AV, Sinitsyn AP. 2009. Isolation and properties of extracellular beta-xylosidases from fungi Aspergillus japonicus and Trichoderma reesei. Biochemistry Moscow. 74:1002–1008.
- Semenova MV, Grishutin SG, Gusakov AV, Okunev ON, Sinitsyn AP. 2003. Isolation and properties of pectinases from the fungus Aspergillus japonicus. Biochemistry Mosc. 68(5):559–569.
- Wang YB, Nguyen TT, Dai YD, Yu H, Zeng WB, Wu CK. 2018. Molecular phylogeny and morphology of Ophiocordyceps unituberculata sp. nov. (Ophiocordycipitaceae), a pathogen of caterpillars (Noctuidae, Lepidoptera) from Yunnan, China. Mycol Prog. 17(6):745–753.
