Abstract
We sequenced the complete plastid genome of Anomodon attenuatus. The genome size is 125,320 bp with two inverted repeat regions of 9,948 bp separated by a large single-copy region of 86,847 bp and a small single-copy region of 18,578 bp. It encodes 114 unique genes including 80 protein-coding genes, 30 tRNA genes and 4 rRNAgenes. The overall GC content is 29.5%, with 26.9, 26.5, and 43.9% in LSC, SSC, IR regions, respectively. 139 SSRs were identified in the genome. Phylogenetic anlysis showed that the position of A.attenuatus is closely related to S.uncinata, another pleurocarpous moss.
Anomodon attenuatus (Hedw.) Hüb. is a widespread moss species distributed over Northern Hemisphere, ranging from North America to Europe and central Asia (Crum et al. Citation1994). Due to their xerophytic feature, A.attenuatus can survive in extreme drought conditions like rocks, tree bases, and calcareous areas (Mahler Citation1980; Crum et al. Citation1994). Genetic background for bryophytes are limited although they may process special desiccation-tolerant mechanism. Here, we assembled and annotated the complete plastid genome of A. attenuatus using the NGS technology. The data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb with accession number CNP0000886.
Anomodon attenuatus was collected from Connecticut (41°49′35″N, 72°10′48″W), USA, and the voucher specimens have been deposited at CONN (collection number BG14206). Genomic DNA (340) was extracted using the modified CTAB method (Sahu et al. Citation2012) and stored at Fairy Lake Botanical Garden, Shenzhen, China. Genome sequencing was performed using BGISEQ500 platform (BGI, Shenzhen, China). A total of 5 Gb high-quality reads were obtained after trimming with SOAPfilter v2.2 using default parameters. Plastid genome was assembled using NOVOPlasty v2.7.3 (Dierckxsens et al. Citation2017) with the plastid genome of Physcomitrella patens (NC_005087.2) plastid as the seed sequence and annotated in CPGAVAS2 (Shi et al. Citation2019) with the plastid genome of Sanionia uncinata (NC_025668.1) as the reference.
The complete plastid genome of A.attenuatus is 125,320 bp in length consisting two inverted repeat regions (IRs) of 9948 bp separated by a large single-copy (LSC) region of 86,847 bp and a small single-copy region (SSC) of 18,578 bp. It encodes 114 unique genes including 80 protein-coding genes, 30 tRNA genes, and 4 rRNAgenes. Among these, 5 tRNA genes (trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and 4 rRNA genes (rrn16, rrn23, rrn4.5, and rrn5) are duplicated in the IR regions. 14 genes (ndhB, ycf66, rpoC1, atpF, ycf3, clpP, rpl2, ndhA, trnA-UGC, trnI-GAU, trnG-UCC, trnL-UAA, trnI-GAU, and trnA-UGC) harbor one intron and 2 genes (trnK-UUU and trnV-UAC) have two introns. The overall GC content of the genome is 29.5%, with 26.9, 26.5, and 43.9% in LSC, SSC, IR regions, respectively.
A total of 139 microsatellite repeats were identified in A. attenuatus plastid genome that can be classified into 84 mononucleotide, 26 dinucleotide, 9 trinucleotide, 15 tetranucleotide, and 5 pentanucleotide using a Perl script MISA (Beier et al. Citation2017). Most microsatellites were found in the LSC region and few in the IR regions, and high A/T ratios were observed in the microsatellites in this genome.
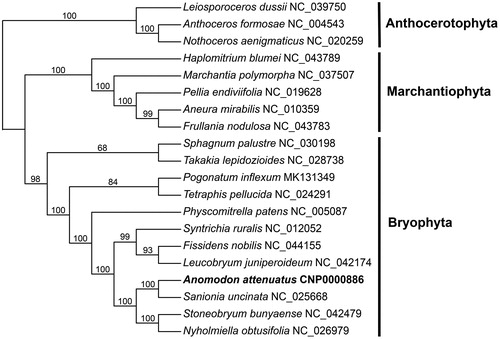
To obtain the phylogenetic position of A.attenuatus within the bryophytes, 20 complete plastid genomes of 20 species, were used to construct a concatenated datasets. Whole plastid genomes were aligned using MAFFT v7.310 (Katoh and Standley Citation2013). The Maximum-Likelihood analysis was performed under the PROTGAMMACPREV model using RAxML v8.2.4 with 500 bootstrap replicates (Stamatakis Citation2014). The phylogenetic tree reveals the position of A.attenuatus is closely related to S.uncinata, and the positions of the other species in the tree are congruent with previous study () (Mizia et al. Citation2019; Park et al. Citation2018).
Acknowledgment
We thank professor Bernard Goffinet for collecting the material of Anomodon attenuatus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Crum H, Sharp A, Eckel P. 1994. The moss flora of Mexico. Memoirs of the New York Botanical Garden. New York Botanical Garden, New York. Vol. 69; p. 1–1113.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mahler WF. 1980. The mosses of Texas: a manual of the moss flora with sketches. Southern Methodist University Herbarium, Dallas, Texas.
- Mizia P, Myszczyński K, Ślipiko M, Krawczyk K, Plášek V, Szczecińska M, Sawicki J, Op Ł. 2019. Comparative plastomes analysis reveals the first infrageneric evolutionary hotspots of Orthotrichum sl (Orthotrichaceae, Bryophyta). Turk J Bot. 43.
- Park M, Park H, Lee H, Lee B-h, Lee J. 2018. The complete plastome sequence of an Antarctic bryophyte Sanionia uncinata (Hedw.) Loeske. IJMS. 19(3):709.
- Sahu SK, Thangaraj M, Kathiresan K. 2012. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol. 2012:1–6.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.