Abstract
Iotaphora admirabilis Oberthür, 1883 belongs to the family Geometridae of Lepidoptera, distributed in many Asia countries such as India, China, Russia, etc. In this study, we described the general features of the complete mitochondrial genome (mitogenome) sequences of this species. The mitogenome is 16,140 bp long and consists of 37 genes (13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes) and one non-coding A + T-rich region, with the typical arrangement found in Lepidoptera. There are 14 genes on the N-strand with the remaining genes on the J-strand. All protein-coding genes start with ATN codons and stop with TAA codons. Phylogenetic analysis, based on the complete mitogenome sequences of 13 species belonging to 3 subfamilies (Larentiinae, Geometrinae, and Ennominae) of Geometridae, shows the sister relationship between Geometrinae and Ennominae, with the placement of Larentiinae as the basal lineage of the two subfamilies.
The Geometridae is one of the most species-rich families of Lepidoptera with almost 21,000 species (Scoble Citation1999). The genera Iotaphora is very unique in Geometridae due to the margins of its fore and hind wings with many radiating lines, which just includes two species in the world, Iotaphora admirabilis Oberthür, 1883 and I. iridicolor Butler, 1880, distributed in China, Russia, India, Nepal, Vietnam, and Myanmar. The larvae of I. admirabilis feed on many plants such as from Salicaceae, Juglandaceae, Bignoniaceae, Betulaceae, and Corylaceae (Han and Xue Citation2011).
The mitochondrial genome (or mitogenome) is a typically double-stranded, circular DNA molecule containing 37 genes (13 protein-coding genes or PCGs, 22 transfer RNA genes or tRNAs, and 2 ribosomal RNA genes or rRNAs) and a non-coding A + T-rich region (Cameron Citation2014), which has been used to study the molecular systematics, population genetics, and molecular evolution (Shao et al. Citation2003; Ma et al. Citation2012; Nelson et al. Citation2012). More mitogenome sequences would be helpful to conduct mitogenome-based phylogeny and to understand the genomic characteristics. Consequently, we sequenced the complete mitogenome of I. admirabilis.
The moths of I. admirabilis were collected in Huangshan Mountains, China (30°04′44.59″ N, 118°08′56.59″ E). The methods of DNA extraction, amplification, and sequencing were same as that described in Li et al. (Citation2018, Citation2019). The specimen (accession number is 20170525004) and the template DNA (accession number is 20170525004DNA) were respectively deposited in the Specimens Room and in the Human and Animal Genetics Laboratory, School of Life Sciences, Huaibei Normal University, China.
The complete mitogenome of I. admirabilis is 16,140 bp long (the GenBank accession number is MK903032), and it consists of 42.5% A, 39.8% T, 10.3% C, and 7.4% G. Like other lepidopterans, the mitogenome of I. admirabilis has 37 genes and one non-coding A + T-rich region (11,280 bp of 13 PCGs, 1476 bp of 22 tRNAs, 2254 bp of 2 rRNAs, and 1005 bp of A + T-rich region). Two rRNAs, 4 PCGs (nad1, nad4, nad4l, nad5), and 8 tRNAs (trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, trnV) are located on the N-strand, the remaining 23 genes are located on the J-strand. All PCGs start with ATN codons and stop with TAA codons. The A + T-rich region of the I. admirabilis mitogenome is located between rrnS and trnM genes, and it has a Lepidoptera-specific poly-T stretch. However, it is obviously longer than the other species of Lepidoptera (Kim et al. Citation2013; Li et al. Citation2018, Citation2019), and the motif sequence is ACATA instead of ATAGA in other geometrid moths.
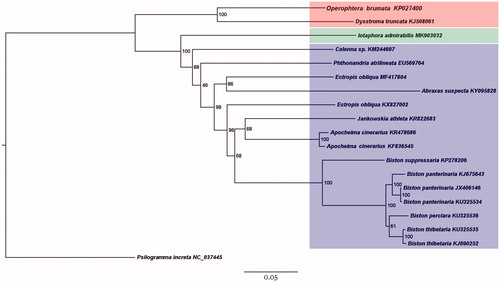
Currently, the complete mitogenome sequences of 13 species belonging to 3 subfamilies (Larentiinae, Geometrinae, and Ennominae) of Geometridae have been verified in GenBank. Phylogenetic analysis based on the nucleotide sequences except A + T-rich regions of these species was performed using maximum-likelihood (ML) method. Psilogramma increta Walker, 1865 (Lepidoptera: Sphingidae) was used as outgroup. The bootstrap value was set 1000 times and the model was GTR + G+I. The result shows the sister relationship between Geometrinae and Ennominae, with the placement of Larentiinae as the basal lineage of the two subfamilies (), which is consistent with Abraham et al. (Citation2001). It is noticeable that two sequences of Ectropis obliqua (MF417804 and KX827002) were not clustered together, which should be the result of the wrong species identification.
Figure 1. Phylogenetic tree for Geometridae, based on 18 mitogenome sequences of 13 species with maximum-likelihood (ML) method. The numbers on the tree are the bootstrap values of ML. The scale bar indicates the number of substitutions per site. GenBank accession numbers of mitogenome sequences are listed after the scientific names of species. The species with red background belong to Larentiinae, green belongs to Geometrinae, and blue belongs to Ennominae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Abraham D, Ryrholm N, Wittzell H, Holloway JD, Scoble MJ, Löfstedt C. 2001. Molecular phylogeny of the subfamilies in Geometridae (Geometroidea: Lepidoptera). Mol Phylogenet Evol. 20(1):65–77.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Han HX, Xue DY. 2011. Fauna of Sinica: insect: Lepidoptera: Geometridae: Geometrinae. Vol. 54. Beijing: Science Press, p. 787.
- Kim MJ, Choi SW, Kim IS. 2013. Complete mitochondrial genome of the larch hawk moth, Sphinx morio (Lepidoptera: Sphingidae). Mitochondrial DNA. 24(6):622–624.
- Li J, Hu KJ, Zhao YQ, Lin RR, Zhang YY, Li Y, Huang ZR, Peng SY, Geng XX, Zhang HJ, Zhang X, et al. 2019. Complete mitogenome of Parum colligata (Lepidoptera: Sphingidae) and its phylogenetic position within the Sphingidae. Zootaxa. 4652(1):126–134.
- Li J, Zhang YY, Hu KJ, Zhao YQ, Lin RR, Li Y, Huang ZR, Zhang X, Geng XX, Ding JH. 2018. Mitochondrial genome characteristics of two Sphingidae insects (Psilogramma increta and Macroglossum stellatarum) and implications for their phylogeny. Int J Biol Macromol. 113:592–600.
- Ma C, Yang P, Jiang F, Chapuis M-P, Shali Y, Sword GA, Kang LE. 2012. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol Ecol. 21(17):4344–4358.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142.
- Scoble MJ. 1999. Geometrid moths of the world: a catalogue (Lepidoptera, Geometridae). Vol. 1, 2. Colingwood: CSIRO, p. xxv + 1016.
- Shao A, Dowton M, Murrell A, Barker SC. 2003. Rates of gene rearrangements and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 20:1612–1619.
