Abstract
Sarcodum scandens is a tropical ornamental liana, distributed in southeastern Asia. The complete chloroplast genome was sequenced using the Illumina Hiseq X-Ten platform. The genome lacks an inverted repeat (IR) region, containing 76 protein-coding genes, 30 tRNAs genes and 4 rRNAs. The overall GC content is 34.1%. A phylogenetic tree based on the whole chloroplast genomes of 14 species indicated that S. scandens belonged to a monophyletic tribe Wisterieae, which was sister to Glycyrrhiza and nested in IRLC group of the subfamily Papilionoideae (Leguminosae).
The tropical liana Sarcodum scandens Lour. (Fabaceae, the legume family) was a species of ornamental plant in southeastern Asia with its showy, purplish-red flowers (Ding Citation1994; Adema Citation1999). The natural distribution of Sarcodum scandens covered the circum-South China Sea region (Geesink Citation1984; Clark Citation2008). Noticeably, this species was recorded in central and southern Hainan island of China (Clark Citation2008; Sun and Pedley Citation2010), but it has been barely witnessed according to our field investigation during the last decade. Few previous study focused on the genome of S. scandens, a good knowledge in genomic information of this species would contribute to the study of population genetics, diversity, gardening and the establishment of efficient protection strategy toward the decreasing natural resource.
The fresh leaves of Sarcodum scandens was collected in Quang Binh, Vietnam (17°29′46″E, 106°19′59″N), and the voucher specimen was deposited in the herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC, collection #: P.K.Loc 11552). We extracted the total genomic DNA with CTAB approach (Doyle Citation1987), the genomic library was prepared and sequenced using the Illumina Hiseq X-Ten platform (Illumina Inc., San Diego, CA, USA). The resultant sequences were filtered following Yao et al. (Citation2016), the adaptor-free reads were then assembled with SPAdes 3.11 (Bankevich et al. Citation2012). We annotated the assembly of complete chloroplast (cp) genome using the Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004) and deposited the genomes in GenBank (accession number: MT039380).
About 1.88 Gb raw reads of S. scandens were obtained, with coverage of 375 × and 132,503 bp in length. The cp genome lacked inverted repeat (IR) region. The genome contained 76 protein-coding genes (CDS), 30 transfer RNA genes (tRNA), 4 ribosomal RNA genes (rRNA), within which 16 genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) had one intron; on the other hand, genes of rps12 and ycf3 had two introns, respectively. Overall GC content of the whole genome was 34.1%.
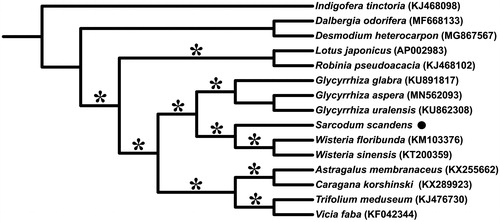
To infer the phylogenetic relationships among this species and its related taxa, whole cp genomes of 13 Papilionoideae species were downloaded from GenBank, which were aligned with that of S. scandens by applying MAFFT v.7 (Katoh and Standley Citation2013). Based on the alignment, a maximum-likelihood (ML) tree was constructed using IQ-TREE v.1.6 (Nguyen et al. Citation2015). The result () showed that the S. scandens, Wisteria floribunda, and W. sinensis constituted the well-supported tribe Wisterieae [as in Compton et al. (Citation2019)], serving as sister of the licorice genus, Glycyrrhiza. This tribe was nested in the inverted repeat-lacking clade (IRLC), which in turn belonged to the clade of Hologalegina as suggested by previous study (Wojciechowski et al. Citation2004; Schrire Citation2005).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adema F. 1999. Notes on Malesian Fabaceae (Leguminosae - Papilionoideae) 5. The genus Sarcodum. Blumea. 44:407–409.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Clark R. 2008. Revision of the genus Sarcodum (Leguminosae: Papilionoideae: Millettieae). Kew Bull. 63(1):155–159.
- Compton JA, Schrire BD, Könyves K, Forest F, Malakasi P, Mattapha S, Sirichamorn Y. 2019. The Callerya Group redefined and Tribe Wisterieae (Fabaceae) emended based on morphology and data from nuclear and chloroplast DNA sequences. PhytoKeys. 125:1–112.
- Ding CS. 1994. Sarcodum. Vol. 40. In: Wei Z, editor, Flora Reipublicae Popularis Sinecae. Beijing: Science Press; pp. 188–189.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Geesink R. 1984. Scala Millettiearum: a survey of the genera of the Millettieae (Legum.-Pap.) with methodological considerations. Leiden: EJ Brill/Leiden University Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Schrire BD. 2005. Millettieae. In: Lewis G, Schrire B, Mackinder B, Lock M, editors. Legumes of the world. Kew: Royal Botanic Gardens.
- Sun H, Pedley L. 2010. Sarcodum. Vol. 10. In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; pp. 174–175.
- Wojciechowski MF, Lavin M, Sanderson MJ. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well‐supported subclades within the family. Am J Bot. 91(11):1846–1862.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6(1):28559.