Abstract
The species Caryopteris incana is a traditional Chinese herbal medicine and ornamental plant. The first complete plastid genome sequences of C. incana reported here was 151,687 bp long, with the large single copy (LSC) region of 83,178 bp, the small single copy (SSC) region of 17,225 bp and two inverted repeats (IRa and IRb) of 25,642 bp. The plastome contained 114 genes, including four ribosomal RNA genes, 30 transfer RNA genes, and 80 protein coding genes. The overall GC content was 38.2%. Result from phylogenetic analysis suggests that C. incana is a member of subfamily Ajugoideae, which is in turn sister to Scutellarioideae and Lamioideae.
The recircumscribed genus Caryopteris Bunge (Cantino et al. Citation1998) comprising seven species, of which C. incana (Thunb. ex Houtt.) Miq. is the most widespread species occurring throughout East Asia. This species is a perennial herb to small woody shrub distribute in China, Japan, and the Korean Peninsula (Ando et al. Citation2015; Xiang et al. Citation2018). In East Asia, this species, due to its anti-inflammatory properties, has been widely used as a traditional medicine for the relief of colds, pertussis, eczema, itchy skin, and venomous snake bites (Mao et al. Citation2016).
Fresh leaves of Caryopteris incana were collected from a living plant cultivated in Kunming Botanical Garden, Yunnan, China (102°44′47.13″E, 25°8′8.09″N). Voucher specimen (Zhao et al. ZF030) was kept in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Total genomic DNA was isolated using the CTAB method (Doyle and Doyle Citation1987) and sequenced on the Illumina HiSeq 2000 Sequencing System at BGI-Shenzhen (Shenzhen, Guangdong, China). The de novo assembling of the chloroplast genome was implemented in the GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle, Jin et al. 2018), and the genome obtained was annotated using software Geneious v.11.0.3 (Kearse et al. Citation2012). The annotated plastid genome sequence has been submitted to GenBank (MN928534).
The whole plastid genome of C. incana was 151,687 bp in length, comprising a pair of inverted repeat regions (IRa and IRb: 25,642 bp) separated by a large single-copy (LSC) region (83,178 bp) and a small single-copy (SSC) region (17,225 bp). The annotated genome comprised 114 genes, including 80 protein-coding genes, four ribosomal RNA genes (rrn16, rrn23, rrn4.5, rrn5), and 30 transfer RNA genes.
Eighteen genes were duplicated in the IR regions, including seven protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf2, ycf15), four ribosomal RNA genes (rrn16, rrn23, rrn4.5, rrn5), and seven transfer RNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC). The overall GC content of C. incana plastid genome is 38.2% (LSC, 36.4%; SSC, 32.3%; IRs, 43.3%).
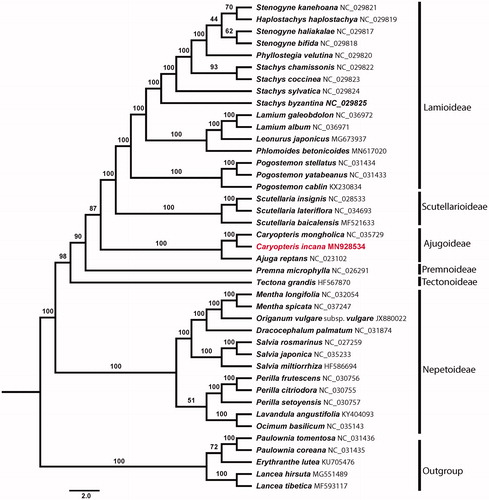
Maximum likelihood (ML) phylogenetic analyses were conducted using RAxML v8.1.11 (Stamatakis Citation2014) as implemented on the on the Cyber infrastructure for Phylogenetic Research (CIPRES) Science Gateway (http://www.phylo.org/, Miller et al. Citation2010), employing the GTR + G model with 1000 bootstrap iterations (-#|-N). Other parameters used the default settings. Phylogenetic analysis based on 80 protein-coding genes of 36 representative plastomes within the family Lamiaceae as in groups. Five species from Phrymaceae (Erythranthe lutea (L.) G.L. Nesom), Paulowniaceae (Paulownia coreana Uyeki, P. tomentosa (Thunb.) Steud.), and Mazaceae (Lancea hirsute Bonati, L. tibetica Hook. f. & Thomson) were selected as outgroup. As a result, C. incana and C. mongholica Bunge form a well-supported clade that is sister to Ajuga reptans L.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ando M, Watanabe H, Matsubara K, Taniguchi A. 2015. Intraspecific phylogenetic relationships of Caryopteris incana in the Tsushima Islands, Japan, using DNA sequence analysis. AJPS. 06(14):2361–2373.
- Cantino PD, Wagstaff ST, Olmstead RG. 1998. Caryopteris (Lamiaceae) and the conflict between phylogenetic and pragmatic considerations in botanical nomenclature. Syst Bot. 23(3):369–386.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 256479. doi:10.1101/256479
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Mao XD, Chou GX, Zhao SM, Zhang CG. 2016. New iridoid glucosides from Caryopteris incana (Thunb.) Miq. and their α-Glucosidase inhibitory activities. Molecules. 21(12):1749.
- Miller MA, Wayne P, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE). IEEE. p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiang CL, Zhao F, Cantino PD, Drew BT, Li B, Liu ED, Soltis DE, Soltis PS, Peng H. 2018. Molecular systematics of Caryopteris (Lamiaceae) and its allies with reference to the molecular phylogeny of subfamily Ajugoideae. taxon. 67(2):376–394.