Abstract
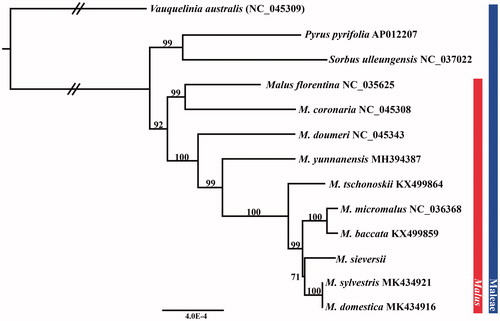
We report a complete chloroplast genome sequence of Malus sieversii. The CP genome was 160,223 bp in length with a typical quadripartite structure. Length of large single-copy (LSC) was 88,334 bp, whereas small single-copy (SSC) was 19,179 bp in length. A pair of inverted repeats (IR) were 26,355 bp. In total, 128 genes were found including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis on concatenated data set retrieved from 78 protein-coding genes revealed that M. sieversii was closely related with both M. ×domestica and M. sylvestris.
Apples (Malus ×domesica Borkh.) are one of the most popular crops around the world and heavily cultivated in temperate regions (Foster et al. Citation2003). Malus sieversii (Lebed.) M. Roem, a wild apple tree, has been recognized as the progenitor of the domesticated apple based on morphological (Robinson et al. Citation2001) and genomic evidence (Coart et al. Citation2006; Velasco et al. Citation2010). Malus sieversii is distributed throughout Central Asia, particularly along the Tian Shan mountain ranges (Lawrence et al. Citation2006). However, the natural habitats of M. sieversii have been disrupted by anthropogenic disturbances, which are resulting in the increased extinction risk as shown in IUCN RED list (Vulnerable). To ensure that the genetic information of the wild apple is preserved for a conservation strategy, we examined the structure of complete chloroplast (CP) genome of M. sieversii.
We collected fresh leaves of a M. sieversii from a natural habitat in Kazakhstan (N 45.506717, E 80.750928). The voucher specimen was prepared and deposited in the Korea National Arboretum Herbarium (KH; Voucher Kz-224). We isolated genomic DNA using the DNeasy Plant Mini Kit followed by the manufacturer’s protocol (Qiagen Inc., Valencia, California, USA). The prepared libraries were sequenced on the MiSeq platform with 550 bp insert sizes. We trimmed the raw reads in Geneious R v. 10.2.2 program (Biomatters Ltd., Auckland, New Zealand). The CP genome was assembled using reference, M. ×domestica (NC_045343) CP genome. Protein-coding genes (including ribosomal RNA) and tRNAs were annotated in Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005), respectively. We inferred the phylogenetic tree of 13 CP genomes in the tribe Maleae including 10 species of Malus based on 78 protein-coding genes (). Based on the assembly results, the CP genome of M. sieversii (MT019538) was 160,223 bp in length with a typical quadripartite structure. Lengths of large single-copy (LSC) and small single-copy (SSC) were 88,334 bp and 19,179 bp, respectively. The inverted repeats (IRa and IRb) were same in length, i.e. 26,355 bp. In total, 128 genes were found, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. According to the inferred phylogeny, Malus formed a monophyletic clade with high statistical support (bootstrap value = 92%). In the phylogeny, M. sieversii, a putative progenitor of the domesticated apple, formed a sister clade with M. ×domestica and M. sylvestris (bootstrap value = 71%). Notably, M. ×domestica appeared to be the most closely related species to M. sylvestris instead of M. sieversii (). The result is somewhat conflicting with the relationship expected from the morphology as M. ×domestica shared the most morphological similarity with M. sieversii (Robinson et al. Citation2001). Given the limited data set available in our study, caution must be taken to interpret the inferred phylogeny. Our findings provide valuable information on the chloroplast genome of the wild apple tree, which can be employed in various botanical studies.
Acknowledgments
We are grateful to several researcher and lab associates for assistance in field collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Coart E, Van Glabeke S, De Loose M, Larsen AS, Roldán‐Ruiz I. 2006. Chloroplast diversity in the genus Malus: new insights into the relationship between the European wild apple (Malus sylvestris (L.) Mill.) and the domesticated apple (Malus domestica Borkh.). Mol Ecol. 15(8):2171–2182.
- Foster T, Johnston R, Seleznyova A. 2003. A morphological and quantitative characterization of early floral development in apple (Malus × domestica Borkh.). Ann Bot. 92(2):199–206.
- Lawrence A, Paudel K, Barnes R, Malla Y. 2006. Adaptive value of participatory biodiversity monitoring in community forestry. Envir Conserv. 33(4):325–334.
- Robinson JP, Harris SA, Juniper BE. 2001. Taxonomy of the genus Malus Mill.(Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Syst E. 226(1–2):35–58.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. 2010. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 42(10):833–839.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.