Abstract
The complete mitochondrial genome of Rhynchoconger ectenurus was sequenced by high-throughput sequencing method. The length of this genome is 17,716 bp, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 large non-coding regions. ND6 and 8 tRNA genes are encoded by L-strand, and others are encoded by H-strand. Phylogenetic tree based on 13 protein-coding genes shows that the clade of Rhynchoconger ectenurus is closely clustered with that of Conger japonicus, and families Anguillidae and Synaphobranchidae have the closer relationship to Congridae, compared to Muraenidae.
Congridae fishes are marine species inhabiting in warm seas. Rhynchoconger ectenurus is a common species in this family and widely distributed in Japan, Korean Peninsula, East China Sea and northern Australia. The specimen was collected from Naozhou island in Zhanjiang, China (geographic coordinate: N20°54′8.42″, E110°46′52.46″). The specimen was preserved in ethanol and registered to the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SW20181071988. This study is the first report of the complete mitogenome of R. ectenurus, and we analyzed its phylogenetic relationships within the order Perciformes.
The complete mitochondrial genome of R. ectenurus was 17,716 bp in length (GenBank accession No. MN256490), including 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 2 control regions (D-Loop). Genes encoding on the genome are similar to other vertebrates that most of these genes are encoded by the H-stand, except for ND6 and eight tRNA genes (tRNA-Gln, -Ala, -Asn, -Cys, -Tyr, -Ser, -Glu, and -Pro) (Shi et al. Citation2015). The gene arrangement is very identical to those in typical fishes (Wang et al. Citation2013), except the ND6 and tRNA-Glu translocated to the position between tRNA-Thr and tRNA-Pro. Overall base composition values for the mitochondrial genome were 32.2, 27.0, 15.8, and 24.9% for A, C, G, and T, respectively, showing slightly AT-bias of 57.1%.
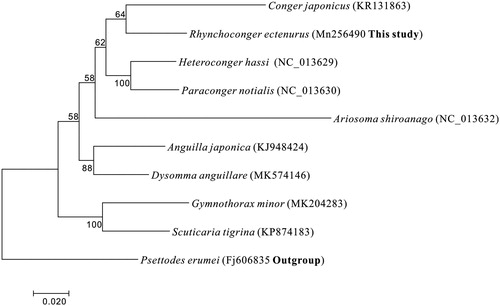
A maximum likelihood (ML) phylogeny tree was constructed using MEGA 7 (Kumar et al. Citation2016), based on first and second codon sequences of 13 protein-coding genes of each mitogenome from nine species in Anguilliformes with Psettodes erumei from Pleuronectiformes as an outgroup (Shi et al. Citation2018). The complete mitochondrial genes of these 10 species are available on GenBank (National Center for Biotechnology Information). In the ML tree, R. ectenurus clustered with Conger japonicus with a good support (64%), Anguilla japonica and Dysomma anguillare formed a clade as a sister lineage to the clade of Congridae family ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Shi W, Chen S, Kong X, Si L, Gong L, Zhang Y, Yu H. 2018. Flatfish monophyly refereed by the relationship of Psettodes in Carangimorphariae. BMC Genom. 19(1):400.
- Shi W, Gong L, Wang SY, Miao XG, Kong XY. 2015. Tandem duplication and random loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes). BMC Genom. 16(1):355.
- Wang Z-M, Shi W, Jiang J-X, Wang S-Y, Miao X-G, Huang L-M, Kong X-Y. 2013. The complete mitochondrial genome of a striped sole Zebrias zebrinus (Pleuronectiformes: Soleidae). Mitochondrial DNA. 24(6):633–635.