Abstract
Aulacophora indica is one of the most important pests threatening the production of melon and vegetables. Samples of A. indica were collected from Zhijiang county and the mitochondrial genome was characterized (GenBank accession number MN162709). The mitogenome consists of a circular DNA molecule of 16,246 bp, with an A/T bias of 78.54%. It comprises 13 protein-coding, 22 tRNA, and 2 rRNA genes. The protein-coding genes have typical ATN (Met) initiation codons and are terminated by typical TAN stop codons.
Aulacophora indica is one of the most important pests threatening the production of melons and vegetables. It is not only harmful to pumpkin, cucumber, loofah, bitter melon, watermelon, melon and other fruits, but also harmful to cruciferous, nightshade, legumes and other vegetables which seriously affect the crop yield and quality. Aulacophora indica adults like eating leaves and petals of melons, damage the pumpkin seedling cortex, bite off the tender stem and eat young fruit. The broken leaves usually formed a circular defect and photosynthesis was inhibited. The larvae feed exclusively on the roots of melons in the ground, which causes wilt stem and dead, and also decays into the adjacent parts of melons, causing rot and loss of edible value (Ahmad et al. Citation2013). However, its mitochondrial genome has not been sequenced completely. Here, we have characterized the complete mitogenome of A. indica to better comprehend its molecular evolution and taxonomic classification (Bai et al. Citation2018).
Adult of A. indica were obtained from Wenan Town (E111.830°, N30.521°), Zhijiang County, Yichang City, Hubei Province, China, on 6 May 2019. The sample was deposited in Guiyang University with Specimen Accession Number: GYU-2019-001. Genomic DNA was isolated and fragmented to build a genomic library of insert size 400 bp that was sequenced (paired end 2 × 150 bp) using an Illumina HiSeq 4000. We obtained 19,884,088 reads of raw data, 16,952,652 of which were high-quality, clean data (96.98%). The genome was assembled de novo with A5-miseq v20150522 (https://github.com/koadman/docker-A5-miseq) (Coil et al. Citation2014) and SPAdes v3.9.0 (http://cab.spbu.ru/software/spades/) (Bankevich et al. Citation2012).
The mitogenome of A. indica consists of a 16,246 bp circular DNA molecule with 40.40% A, 38.06% T, 12.81% C, and 8.63% G, which is an A/T bias of 78.54%. The AT- and GC-skews of the major strands of the mitogenome were calculated to be approximately 0.031 and −0.195, respectively.
The mitogenome of A. indica contains 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. All 13 PCGs have typical ATN (Met) start codons: nad1 and cox3 – ATA; nad2, nad5, nad6, cox1, cox2, and atp8 – ATT; cox3, atp6, nad3, nad4, nad4l and cox – ATG. Twelve genes (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4l, nad6, cob) have a TAA stop codon; nad1 has a TAG stop codon. These genomic features of A. indica are similar to the mitochondrial genome of the species A. lewisii of the same genus (Song et al. Citation2018).
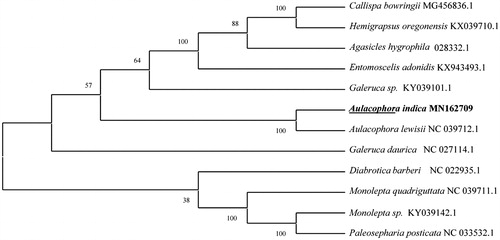
To validate the phylogenetic position of A. indica, the mitogenome DNA sequences from 11 species of Chrysomeloidea beetles were used to construct a phylogenetic tree by the maximum likelihood method using the MEGA 7 software (Kumar et al. Citation2016; Yuyu et al. Citation2019) (). Overall, our study provides insight into the mitogenome of A. indica, which will be useful for its taxonomic classification and further phylogenetic reconstruction.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ahmad W, Naeem M, Bodlah I. 2013. Genus Aulacophora Chevrolat, 1836 (Coleoptera: Chrysomelidae) from Pothohar, Punjab, Pakistan. Pak J Zool. 45(3):868–871.
- Bai Y, Li C, Yang M, Liang S. 2018. Complete mitochondrial genome of the dark mealworm Tenebrio obscurus Fabricius (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA Part B. 3(1):171–172.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski A. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Coil D, Jospin G, Darling A. 2014. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Kumar S, Stecher G, Tamura K. evolution. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Song N, Yin X, Zhao X, Chen J, Yin J. 2018. Reconstruction of mitogenomes by NGS and phylogenetic implications for leaf beetles. Mitochondrial DNA Part A. 29(7):1041–1050.
- Yuyu W, Liming W, Juan Y, Xianquan Q. 2019. The complete mitochondrial genome of Monolepta occifuvis Gressitt Kimoto (Coleoptera: Chrysomelidae:Galerucinae). Mitochondrial DNA Part B. 4(1):1654–1655.