Abstract
Borago officinalis is a kind of vegetable and traditional medicinal herb in the Mediterranean region, and now it has spread many areas around the world. In this study, we sequenced a complete chloroplast (cp) genome sequence of B. officinalis to investigate its phylogenetic relationship with other species. The whole cp genome of B. officinalis is 149,835 bp in length with 37.82% overall GC content, including a large single-copy (LSC) region of 78,840 bp and a small single-copy (SSC) region of 16,967 bp, which are separated by a pair of inverted repeats (IRs) of 27,014 bp. The cp genome contains 110 genes, including 76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis based on cp genome sequences showed that B. officinalis is closely related to Antirhea chinensis, Mitragyna speciosa and Neolamarckia cadamba.
Borago officinalis Linn. is a plant belonging to the family Boraginaceae. It is an annual herb native to the Mediterranean region of Europe, but now cultivated in many areas of the world (Montaner et al. Citation2000). It is currently used as a vegetable. The petioles, leaves, and stems are eaten raw in salads or fried lightly, without affecting the nutritional attributes of the plant. Flowers can be made into candies, preserved or added to salads and juice drinks. It is also used widely in traditional medicine. Its leaves can be used as a demulcent, diuretic, expectorant or emollient (Francisco et al. Citation2014) and its seeds can be used in pharmaceutical products because of the high γ-linolenic acid (GLA, cis-6,9,12-octadecatrienoic acid, 18:3Δ6,9,12) (Mercedesdel et al. Citation2008) content in seed oil. So far, no studies on the genome of B. officinalis have been published. Here, we reported the complete chloroplast (cp) genome sequence of B. officinalis, as well as reconstructed its phylogenetic tree with other groups based on cp genomes.
Plant materials were collected from the greenhouse, Shandong Academy of Agricultural Sciences, Shandong province of China, located at 36°42′23″N, 117°4′25″E; meanwhile, a voucher specimen (No.2018-Bo06) was collected and deposited at Shandong Center of Crop Germplasm Resources. Total genomic DNA was extracted using the CTAB method and sequenced with Illumina Hiseq 4000 (Illumina, San Diego, CA) platform. The raw data were filtered using Trimmomatic v.0.32 (Bolger et al. Citation2014) with default settings. Then, pair-end reads of clean data were assembled into a circular contig using GetOrganelle.py (Jin et al. Citation2018). Finally, the cpDNA was annotated using GeSeq (Michael et al. Citation2017) and tRNAscan-SE (Lowe and Chan Citation2016).
The annotated chloroplast genome was submitted to the GenBank under the accession number MN823729. The whole cp genome sequence is 149,835bp in length with a typical circular quadripartite structure. A pair of inverted repeats regions (IRa and IRb) of 27,014 bp are separated by a large single-copy (LSC) region of 78,840 bp and a small single-copy (SSC) region of 16,967 bp. The overall GC content of cp genome is 37.82%. The cp genome contains 110 genes, including 76 protein-coding genes, 4 rRNA genes, 30 tRNA genes, and 4 genes have been inferred to be pseudogenes. Among these genes, 20 genes are duplicated in the inverted repeat regions, 12 genes, and 6 tRNA genes contain one intron, whereas two genes (clpP and ycf3) have two introns.
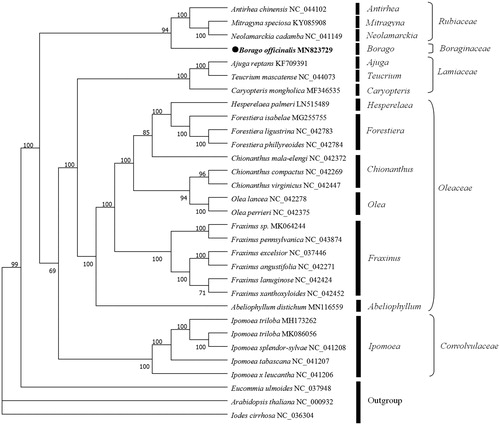
A total of 30 cp genome sequences were downloaded from the NCBI database to confirm the phylogenetic position of B. officinalis. The sequences were aligned using MAFFT v7 (Katoh and Standley Citation2013), and ML tree inference was implemented in IQTREE 1.6.12 (Nguyen et al. Citation2015) with 1000 bootstrap replicates. The results showed that B. officinalis is closely related to Antirhea chinensis, Mitragyna speciosa and Neolamarckia cadamba (). The complete chloroplast genome of B. officinalis would lay a solid foundation for further phylogenetic studies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Francisco S, Bryshila L, Sara P, Gordon MH, Pilar AM. 2014. Extraction of antioxidants from Borage (Borago officinalis L.) leaves—optimization by response surface method and application in oil-in-water emulsions. Antioxidants. 3:339–357.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. doi: 10.1101/256479
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mercedesdel R, Rafael F, Antonio HB. 2008. Distribution of fatty acids in edible organs and seed fractions of borage (Borago officinalis L.). J Sci Food Agr. 88:248–255.
- Michael T, Pascal L, Tommaso P, Elena SUJ, Axel F, Ralph B, Stephan G. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. W1(45):W6–W11.
- Montaner C, Floris E, Alvarez JM. 2000. Is self-compatibility the main breeding system in borage (Borago officinalis L.)? Theor Appl Genet. 101(1-2):185–189.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.