Abstract
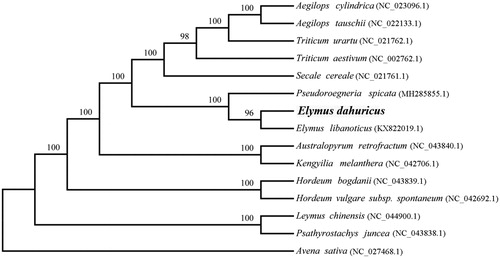
Elymus dahuricus, a widely distributed perennial herb on the Qinghai-Tibetan Plateau, is highly adapted to alpine regions. In this study, we first reported the complete chloroplast (cp) genome of E. dahuricus using Illumina sequencing data. The size of the chloroplast genome is 135,096 bp, including a pair of inverted repeat regions (IRs, 20,812 bp), a large single copy region (LSC, 80,701 bp), and a small single single copy region (SSC, 12,771 bp). The chloroplast genome of E. dahuricus contained 111 genes, including 78 protein-coding genes, four ribosomal RNA genes, and 29 transfer RNA genes. Phylogenetic tree analysis suggested that E. dahuricus was closely related to E. libanoticus.
Elymus dahuricus Turcz. ex Griseb., a perennial herb species in Triticeae, is widely distributed in Central Asia, Russia, Mongolia, China, Korea, Japan, and Northern India (Baum et al. Citation2011). It can resistant extreme winter temperatures and adverse soil conditions, and is one of the indicator species in the Steppe, abandoned croplands, and yardangs (Kawada et al. Citation2011). E. dahuricus is used as a high-quality forage and is a common species that improves degraded grassland, especially in Qinghai-Tibetan Plateau below 3000 m (Cui et al. Citation2019). In addition, no visible symptoms of stem rust and loose smut were found in E. dahuricus, which may be a potential candidate for screening biotic tolerance (Pradheep et al. Citation2019). In this study, we assembled the complete cp genome of E. dahuricus (Genbank accession number: MN895932) to provide genomic and genetic sources for further research.
The seeds of E. dahuricus were collected from Gonghe (36°35′N, 100°29′E), Qinghai Province, China. The voucher specimen was deposited in Herbarium of the Northwest Institute of Plateau Biology (HNWP, No. LRJ201814), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Total genomic DNA was extracted from laboratory-grown seedlings leaves with the modified CTAB method and quantified (Allen et al. Citation2006). Sequencing was performed with an Illumina HiSeq 2500 Platform (Wuhan Benagen Tech Solutions Company Limited, Wuhan, China), and 10.83 GB of clean data was generated. Then the clean reads were assembled into circular contigs using SPAdes Version 3.10.1(Bankevich et al. Citation2012). Finally, the cpDNA was annotated using the web application GeSeq (Michael et al. Citation2017).
The complete cp genome of E. dahuricus is 135,096 bp in length. It exhibited a typical circular and quadripartite structure including two copies of inverted repeats (IRs, 20,812 bp) separated by the large single copy (LSC, 80,701 bp) and small single copy (SSC, 12,771 bp) regions. Overall, GC contents of the cp genomes were 38.36%. A total of 111 genes were annotated, including 78 protein-coding genes (PCGs), 29 transfer RNA genes (tRNA), and four ribosomal RNA genes (rRNA). A phylogenetic analysis was performed using cp genome sequences of E. dahuricus and other 13 species in Triticeae, with Avena sativa as outgroup. MAFFT version 7.017 and the web application CIPRES were used to construct a maximum likelihood tree (Miller et al. Citation2010; Kazutaka and Standley Citation2013). The MP tree indicated that E. dahuricus is closely related to E. libanoticus with a high support rate (). This study was the first report on complete cp genome of E. dahuricus which would be beneficial to potential studies on phylogenetics of the genus and related group in Triticeae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Baum BR, Yang JL, Yen C, Agafonov AV. 2011. A taxonomic synopsis of the genus Campeiostachys Drobov. J Systemat Evol. 49(2):146–159.
- Cui GW, Ji GX, Liu SY, Li B, Lian L, He WH, Zhang P. 2019. Physiological adaptations of Elymus dahuricus to high altitude on the Qinghai-Tibetan Plateau. Acta Physiol Plant. 41(7):115.
- Kawada K, Wu YN, Nakamura T. 2011. Land degradation of abandoned croplands in the Xilingol steppe region, Inner Mongolia, China. Grassland Sci. 57(1):58–64.
- Kazutaka K, Standley M. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, Ulbricht-Jones ES, Axel F, Ralph B, Stephan G. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6-W11.
- Miller MA, Pfeiffer WT, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010. Gateway Computing Environments Workshop (GCE), IEEE Computer Society. New Orleans, LA, pp. 1–8
- Pradheep K, Singh M, Sultan SM, Singh K, Parimalan R, Ahlawat SP. 2019. Diversity in wild relatives of wheat: an expedition collection from cold-arid Indian Himalayas. Genet Resour Crop Evol. 66(1):275–285.