Abstract
The complete chloroplast genome of Eriocaulon henryanum is presented in this study. Total chloroplast genome is 151,051 bp in length, containing a large single-copy region (LSC) of 81,171 bp and a small single-copy region (SSC) of 16,978 bp, separated by a pair of inverted repeats (IRS) of 26,451 bp. A total of 114 genes, including 80 protein-coding genes, 30 tRNA genes, and 4 ribosomal RNA genes. The overall A + T content is 64.2%. Phylogenetic relationship analysis shows that Eriocaulon henryanum was relatively close to Eriocaulon nepalense.
Eriocaulon henryanum is a kind of grass that belongs to the family of Eriocaulaceae (Ma Citation1997). It grows in the damp soil of the valley and mainly distributed in China, Thailand, and Vietnam (Zhang Citation1999, Ma et al. Citation2000). The male corolla middle-lobe of E. henryanum extends out of the floral bract and sepals obviously, which is taken as a key identification characteristic of this species. The chloroplast genome-scale data have proven to be useful in resolving phylogenetic relationships (Yang et al. Citation2016). In this study, we report the complete chloroplast genome sequence of E. henryanum.
The specimen of E. henryanum was collected from Huadianba, Yunnan Province, China (100°00′50.63″E, 25°52′15.09″N) and stored at the Herbarium of Beijing Forestry University with an accession number DH-010. Total DNA was extracted using CTAB method (Doyle and Doyle Citation1987). Then, an Illumina HiSeq 4000 platform at BioMarker (http://www.biomarker.com.cn/, China) was applied to perform 2 × 150 bp pair-end sequencing. Clean reads were mapped to the published chloroplast genome of E. decemflorum as references (Darshetkar et al. Citation2019) using map function of Geneious R11 (Kearse et al. Citation2012). Filtered reads were then used for de novo assembly with Geneious R11. Gaps were bridged using fine tuning function of Geneious R11. The complete chloroplast sequence was annotated using Plann (Huang and Cronk Citation2015).
The complete chloroplast sequence of E. henryanum is 151,051 bp, including the large single-copy region (LSC, 81,171 bp), the small single-copy region (SSC, 16,978 bp), and a pair of 26451 bp inverted repeat regions (IRs). The circular genome contains 114 genes, including 80 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. The overall A + T content of the circular genome was 62.4%.
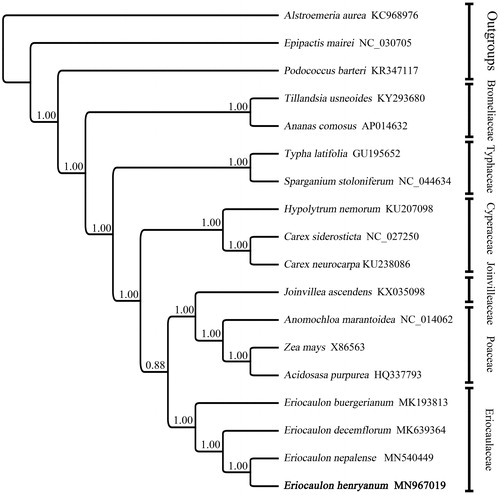
Available complete chloroplast genome sequences of Eriocaulaceae and other families in the Poales were downloaded from GenBank. Bayesian inference analyses (Ronquist and Huelsenbeck Citation2003) was conducted for phylogeny reconstruction (). As shown in the phylogenetic tree, E. henryanum is closely related to E. nepalense compared with other species of Eriocaulon.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK. 2019. Understanding evolution in Poales: Insights from Eriocaulaceae plastome. PLoS ONE. 14(8):e0221423.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Ma WL. 1997. Eriocaulaceae. In: Wu KF, editor. Flora Reipublicae Popularis Sinicae. Vol. 13. Beijing: Science Press; pp. 11–12.
- Ma WL, Zhang ZX, Thomas S. 2000. Eriocaulon nepalense Prescott ex Bongard. in: Wu ZY & Raven P, editor. Flora of China. Vol. 24. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; pp. 11–12.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 07:959.
- Zhang ZX. 1999. Monographie der Gattung Eriocaulon in Ostasien. Dissertationes Botanicae. 313:140–143.