Abstract
Here, we determined the complete mitogenome sequence of Euspira gilva (Littorinimorpha: Naticidae). The complete mitochondrial genome is circular and 16,119 bp in length, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and 22 transfer RNA genes. All PCGs and rRNA genes are encoded on the heavy strand while tRNA genes are distributed in both the strands. The overall nucleotides base composition of the heavy strand is A (31.80%), G (14.72%), C (13.72%), and T (39.76%), with an A + T bias (71.56%). All of the PCGs had ATG as their start codon. The common termination codon is TAA, except for nd4l terminated with TAG. The 22 tRNA genes could be folded into a typical clover-leaf secondary structure. As seen from the phylogenetic tree, E. gilva has a more close relationship with Neverita didyma and Glossaulax reiniana.
Euspira gilva, belonging to the family Naticidae (Gastropoda: Littorinimorpha), is a benthic species widely distributed along the coast of China, Japan and Korea (Zhang Citation2016). Due to its high nutritive and economic value, E. gilva is one of the most commercially important resources for the fishermen communities. Similar to Neverita didyma, the populations of E. gilva is decreasing every year due to over-harvesting (Liu et al. Citation2013; Zhao et al. Citation2018). Previous studies have focused on the morphology and distribution (Leng et al. Citation2013; Zhang Citation2016; Huang et al. Citation2017), but genetic studies on E. gilva are limited. Partial sequences of E. gilva mitogenome have been determined as part of phylogenetic studies of caenogastropod (Zou et al. Citation2011; Sun et al. Citation2012); however, to date, there is no report of complete sequence and characterization of mitochondrial chromosome for E. gilva.
In this paper, we present the complete mitochondrial genome of E. gilva and constructed the phylogenetic relationship of 21 Littorinimorpha species, which, we expect could be valuable for studying population genetics and helpful for rational utilization of this species. The specimen was collected from Laizhou Bay (37.78°N, 119.33°E), China, during April 2019. Specimen (voucher no. FIO-Y-2019-0017) was deposited in the Biodiversity Lab of the First Institute of Oceanography, MNR. The complete mitochondrial genome of E. gilva (GenBank accession no. MN419026) is 16,119 bp in length, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (12SrRNA and 16S rRNA), 22 transfer RNA genes (tRNA), which is the same as the other Naticidae species, N. didyma (Wang et al. Citation2019) and Glossaulax reiniana (Li et al. Citation2018). The PCGs sequences correspond to the genes cox1, cox2, cox3, cob, atp6, atp8, nd1, nd2, nd3, nd4, nd4l, nd5, nd6. All PCGs and rRNA genes encoded on the heavy strand, while the tRNA genes are distributed in both the strands. The overall nucleotides base composition of the heavy strand is A (31.80%), G (14.72%), C (13.72%), and T (39.76%), with an A + T bias (71.56%). All the PCGs begin with an ATG codon. The common termination codon is TAA, except for nd4l terminated with TAG. The longest gene is nd5 (1719 bp) among the PCGs, whereas the shortest is atp8 (159 bp). The 2 rRNA genes, 12S rRNA (953 bp) and 16S rRNA (1366 bp) are located between the tRNA-Glu(TTG) and tRNA-Leu(TAA) genes and are separated by the tRNA-Val(TAC) gene. The 22 tRNA genes vary from 62 to 72 bp in length, and could be folded into the typical cloverleaf secondary structure.
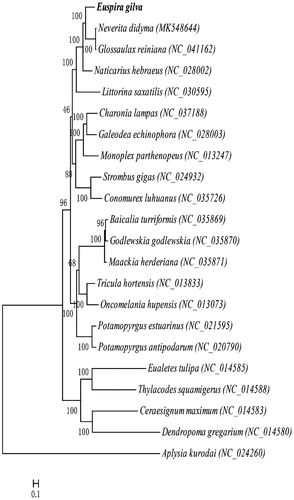
To explore the phylogenetic relationship of Littorinimorpha and the status of E. gilva, we constructed a phylogenetic tree based on the maximum likelihood (by RAxML 8.1.5, Heidelberg, Germany) (Alexandros Citation2006) analysis using the 13 PCGs of mitochondrial genomes of E. gilva and the other 20 species of Littorinimorpha, Aplysia kurodai was used as the outgroup (). According to the phylogenetic analysis, E. gilva has a more close relationship with N. didyma and G. reiniana from the same family.
Figure 1. Maximum likelihood tree based on PCGs of mitochondrial genomes of Euspira gilva (MN419026) and Baicalia turriformis (NC_035869), Godlewskia godlewskia (NC_035870), Maackia herderiana (NC_035871), Galeodea echinophora (NC_028003), Potamopyrgus antipodarum (NC_020790), Potamopyrgus estuarinus (NC_021595), Littorina saxatilis (NC_030595), Glossaulax reiniana (NC_041162), Naticarius hebraeus (NC_028002), Tricula hortensis (NC_013833), Oncomelania hupensis (NC_013073), Charonia lampas (NC_037188), Monoplex parthenopeus (NC_013247), Conomurex luhuanus (NC_035726), Strombus gigas (NC_024932), Ceraesignum maximum (NC_014583), Dendropoma gregarium (NC_014580), Eualetes tulipa (NC_014585), Thylacodes squamigerus (NC_014588), Neverita didyma (MK548644) and Aplysia kurodai (NC_0242603) is used as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alexandros S. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Huang XK, Xiao GQ, Zhang P, Zhang YP, Wang TG, Zhong W, Zhang LN, Lin SZ. 2017. The relationship between morphometric traits and body weight of Lunatica gilva. J Aquacult. 38:25–29.
- Leng Y, Zhang JM, Liu S, Liu XD, Zhang HL, Zhang AJ. 2013. Marine biodiversity in the Yellow River Estuary and Adjacent Water. Qingdao: China Ocean University Press; p. 253.
- Li PY, Yang Y, Li YG, Sun SE. 2018. The complete mitochondrial genome of Glossaulax reiniana (Littorinimorpha: Naticidae). Mitochondrial DNA Part B. 3(2):1263–1264.
- Liu H, Xu M, Wu C. 2013. Evaluation of nutritional composition in Nevertia didyma and Natica vitellus from Zhoushan Sea area. Food Sci. 34:228–231.
- Sun Y, Li Q, Kong LF, Zheng XD. 2012. DNA barcoding of Caenogastropoda along coast of China based on the COI gene. Mol Ecol Resour. 12(2):209–218.
- Wang ZX, Liu CX, Liu HZ, Wang B, Pang M, Zheng FR. 2019. The mitochondrial genome of the marine gastropod Neverita didyma (Roding, 1798) (Mollusca: Gastropoda). Mitochondrial DNA Part B. 4 (1):1545–1546.
- Zhang SP. 2016. Fauna sinica. Invertebrata 56. Mollusca: Gastropoda: Strombacea and Naticacea. Beijing: Science Press.
- Zhao D, Kong L, Yu H, Li Q. 2018. Cryptic genetic diversity of Neverita didyma in the coast of China revealed by phylogeographic analysis: implications for management and conservation. Conserv GenetGenet. 19(2):275–282.
- Zou SM, Li Q, Kong LF. 2011. Additional gene data and increased sampling give new insights into the phylogenetic relationships of Neogastropoda, within the caenogastropod phylogenetic framework. Mol Phylogenet Evol. 61(2):425–435.
