Abstract
In this study, complete mitochondrial genome of the Lista haraldusalis (Walker, 1859) is sequenced through next-generation sequencing method. The L. haraldusalis mitogenome is a circular, double-stranded molecule, with 15,213 bp in length. The typical 37 mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and an A + T-rich region are recognized. Gene content and arrangement are highly conserved and typical of Lepidoptera. Interestingly, in the intergenic region between trnN and trnS1, a microsatellite sequence of (TA)12 is recognized. Phylogenetic analysis based on the 13 PCGs plus two rRNAs sequences recovered that the Pyralidae and Crambidae are reciprocally monophyletic. The L. haraldusalis sequenced in this study is nested into the Pyralidae clade, reinforcing previous morphological and molecular studies.
Mitochondrial genomes (mitogenomes) contain high genetic information and have been demonstrated to be effective to facilitate evolutionary studies for many animals including insects (Timmermans et al. Citation2014). The Pyraloidea, with approximately 15,500 described species worldwide, is one of the largest superfamilies in Lepidoptera (van Nieukerken et al. Citation2011). Up to now, mitochondrial genomes for only 37 pyraloid species have been reported. In this study, the next-generation sequencing (NGS) method is employed to obtain the complete mitogenome of a pyraloid species, Lista haraldusalis (Walker, 1859). This species, belonging to the Pyralidae, is widely distributed in China.
Adult specimens were collected from Mountain range Jigongshan (114°06′56″E, 31°49′25″N), Henan Province, China. After species identification, the genomic DNA was extracted for library construction. An Illumina Miseq platform was used for sequencing with the strategy of 150 paired-ends. Voucher specimens are deposited in the Biology Laboratory of Zhoukou Normal University (accession number: 2019JGSA18), China.
The L. haraldusalis mitogenome (GenBank accession number: MT081380) is a circular, double-stranded molecule, with 15,213 bp long. The typical 37 mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs) plus an A + T-rich region in Lepidoptera are present. Gene content and arrangement are highly conserved and typical of Lepidoptera. The nucleotide composition is A 40.5%, G 7.7%, C 10.8%, and T 41.1%, showing a highly A/T bias as commonly present in insects (Boore Citation1999).
The total length of 13 PCGs of P. chloerata is 11,223 bp, encoding 3740 amino acids. The standard ATN is used as a start codon for most PCGs, with an exception CGA for the cox1. TAA is routinely used as a stop codon, but the incomplete termination codon T is recognized in cox2, nad6, and nad4. All 22 tRNAs exhibit a typical clover-leaf secondary structure, but trnS1 (AGN) lacks the DHU arm, a common feature in Lepidoptera insects (Garey and Wolstenholme Citation1989). Two rRNA genes, rrnS and rrnL were recognized with the lengths 786 bp and 1335 bp, respectively. Nine gene overlapping regions were found, with the lengths ranging from 1 to 17 bp. Among the 13 intergenic regions, a large one (49 bp) between trnQ and nad2 was recognized and this sequence is routinely regarded as an autapomorphy of Lepidoptera (Cao et al. Citation2014). Besides, the intergenic region (23 bp) between trnS2 and nad1 was also recognized and a typical feature for this region is the presence of the motif ‘ATACTAA’. Interestingly, in the intergenic region between trnN and trnS1, a microsatellite sequence of (TA)12 was recognized. As the largest non-coding region, the A + T-rich region is 310 bp long and contains typical conserved sequence elements such as the motif ‘ATAGA’ and subsequent poly-T structure.
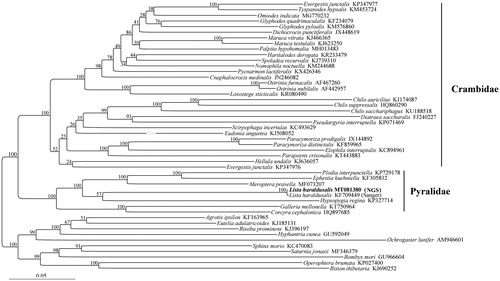
Phylogenetic trees were constructed based on a dataset consisting of 13 PCGs and two rRNAs sequences from the L. haraldusalis and other pyraloid species with mitogenome available. Maximum likelihood analysis () consistently recovered that the Pyralidae and Crambidae are reciprocally monophyletic with the highest supports. The L. haraldusalis we sequenced, together with the same species sequenced by Sanger method as reported in (Ye et al. Citation2015), are nested into the Pyralidae clade.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cao SS, Yu WW, Sun M, Du YZ. 2014. Characterization of the complete mitochondrial genome of Tryporyza incertulas, in comparison with seven other Pyraloidea moths. Gene. 533:356–365.
- Garey JR, Wolstenholme DR. 1989. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA(serAGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons . J Mol Evol. 28(5):374–387.
- Timmermans M, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet E. 79:169–178.
- van Nieukerken EJ, Kaila L, Kitching IJ, Kristensen NP, Lees DC, Minet J, Mitter C, Mutanen M, Regier JC, Simonsen TJ, et al. 2011. Order Lepidoptera Linnaeus, 1758. Zootaxa. 3148(1):212–221.
- Ye F, Yu HL, Li PF, You P. 2015. The complete mitochondrial genome of Lista haraldusalis (Lepidoptera: Pyralidae). Mitochondr DNA. 26(6):853–854.