Abstract
Hipparchia autonoe (Esper, 1783) is a protected butterfly species found in Mt. Halla in South Korea. We have determined mitochondrial genome of H. autonoe collected in Mt. Halla. The circular mitogenome of H. autonoe is 15,300 bp long, which is shorter than previously sequenced mitogenome by 189 bp due to differences of tandem repeats. It includes 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNAs. The base composition was AT-biased (78.9%). Nineteen single nucleotide polymorphisms and one insertion and deletion were identified between the two individuals of H. autonoe captured in Mt. Halla, presenting enough genetic diversity of H. autonoe within population.
Hipparchia autonoe (Esper, 1783) belonging to Nymphalidae family is Palearctic butterfly species, distributed from Korea to the Caucasus (Gorbunov Citation2001). In Korean peninsula, H. autonoe is restricted to only some areas including Mt. Halla (>1,300 m altitudes) and alpine regions of northern Korean peninsula (Cho et al. Citation2013). The isolated population in Mt. Halla has been regarded as the remnants of the Pleistocene glaciations when Jeju island was connected to Korean peninsula (Joo and Kim Citation2002). Due to global warming and its small population, H. autonoe is endangered in South Korea, thus was designated as natural monument No. 458 and is listed as first-degree endangered wild animal (Cho et al. Citation2013).
To investigate genetic diversity of H. autonoe within population, we completed its mitogenome from the sample collected in Mt. Halla, Korea (37°45′74″N, 126°94′84″E; the specimen in InfoBoss Cyber Herbarium (IN); INH-00023). DNA was extracted using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeqX (Macrogen, Korea) were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its mitogenome based on previous H. autonoe mitogenome (NC_024581; Kim et al. Citation2010).
H. autonoe mitogenome (GenBank accession is MT090762) is 15,300 bp long, shorter than former mitogenome (NC_024581) by 189 bp due to decrease of tandem repeats in control region. It contains 13 protein-coding genes (PCGs), 22 tRNAs, and 2 rRNAs. The base composition was AT-biased (78.9%) and gene order was identical to other Nymphalid mitogenomes.
Nineteen single nucleotide polymorphisms (SNPs) and a single insertion and deletion (INDEL) were found by comparing two H. autonoe mitogenomes, which were less than those of other insect species (Choi et al. Citation2019; Park, Kwon, et al. Citation2019; Park et al. Citation2019; Park et al. Citation2019; Seo, Jung, et al. Citation2019; Seo, Lee, et al. Citation2019). However, they were relatively diverse considering its small population. Sixteen of 19 SNPs (84.2%) were placed within PCGs, two were in 16S rRNA gene, and one was in the intergenic region between CYTB and trnS2. Eleven synonymous SNPs change third bases of each codon; while 3 and 2 non-synonymous SNPs were in the first and second bases, respectively, affecting COX3, ND5, CYTB, and ND1. One transversion was found in the third base of the last codon of ND1, not affecting translational product due to post-transcriptional modifications.
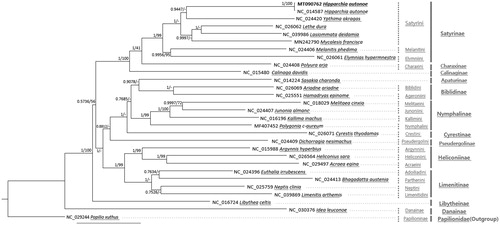
We inferred the phylogenetic relationship based on 29 Nymphalidae mitogenomes including two H. autonoe mitogenomes and one outgroup species. Concatenated multiple sequence alignments of 13 PCGs by MAFFT 7.450 (Katoh and Standley Citation2013) were used for constructing bootstrapped maximum likelihood and Bayesian inference phylogenetic trees with MEGA X (Kumar et al. Citation2018) and Mr. Bayes (Huelsenbeck and Ronquist Citation2001), respectively. Phylogenetic trees were overall congruent to the previous studies (Wu et al. Citation2014; Espeland et al. Citation2018) except that (i) Danainae, not Libytheinae, was sister to all other clades and (ii) Nymphalinae did not cluster with Cyrestinae ().
Figure 1. Bayesian inference (1,000,000 generations) and maximum likelihood (1,000 bootstrap repeats) phylogenetic trees based on 28 Nymphalidae mitochondrial genomes: Hipparchia autonoe (MT090762 in this study and NC_014587), Lethe dura (NC_026062), Ypthima akragas (NC_024420), Lasiommata deidamia (NC_039986), Mycalesis francisca (MN242790), Melanitis phedima (NC_024406), Elymnias hypermnestra (NC_026061), Polyura arja (NC_024408), Calinaga davidis (NC_015480), Sasakia charonda (NC_014224), Ariadne ariadne (NC_026069), Hamadryas epinome (NC_025551), Melitaea cinxia (NC_018029), Junonia almana (NC_024407), Kallima inachus (NC_016196), Polygonia c-aureum (MF407452), Cyrestis thyodamas (NC_026071), Dichorragia nesimachus (NC_024409), Argynnis hyperbium (NC_015988), Heliconius sara (NC_026564), Acraea egina (NC_029497), Euthalia irrubescens (NC_024396), Bhagadatta austenia (NC_024413), Neptis clinia (NC_025759), Limenitis arthemis (NC_039869), Libythea celtis (NC_016724), Idea leuconoe (NC_030376), and one Papilionidae species, Papilio xuthus (NC_029244) as an outgroup. Phylogenetic tree was drawn based on Bayesian inference tree. The numbers above branches indicate posterior probability of Bayesian inference tree and bootstrap support value of maximum likelihood phylogenetic tree, respectively. Tribe names are displayed as light gray color and subfamily names were written as dark gray color.
Disclosure statement
No conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cho Y, Park JS, Kim MJ, Choi DS, Nam SH, Kim I. 2013. Genetic relationships between Mt. H alla and Mongolian populations of Hipparchia autonoe (Lepidoptera: Nymphalidae). Entomol Res. 43(3):183–192.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondr DNA B. 4(1):1677–1678.
- Espeland M, Breinholt J, Willmott KR, Warren AD, Vila R, Toussaint EFA, Maunsell SC, Aduse-Poku K, Talavera G, Eastwood R, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.
- Gorbunov P. 2001. The butterflies of Russia: classification, genitalia, keys for identification. [Thesis]. Ekaterinberg.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Joo H, Kim S. 2002. Butterflies of Jeju Island. Junghaengsa Co.185.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim MJ, Wan X, Kim K, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of Endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Af J Biotechnol. 9(5):735–754.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, 1000 Genome Project Data Processing Subgroup, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondr DNA B. 4(1):1674–1676.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondr DNA B. 4(2):3007–3009.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863)(Lepidoptera: Crambidae). Mitochondr DNA B. 4(1):850–851.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondr DNA B. 4(2):2242–2243.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondr DNA B. 4(2):3918–3920.
- Wu L-W, Lin L-H, Lees DC, Hsu Y-F. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genom. 15(1):468.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genom Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics BioMed Central. 12(Suppl 14):S2.
