Abstract
Rheum nobile is a model species for investigating alpine adaptation in ‘glasshouse’ plants. In this study, the complete chloroplast (cp) genome of R. nobile was assembled based on next-generation sequencing. The cp genome was 161,250 bp in length, including a large single copy (LSC) region of 86,290 bp, a small single copy (SSC) region of 12,838 bp and a pair of inverted repeats (IRs), each of 31,061 bp. The genome contained 127 genes, including 82 protein-coding genes, 35 tRNA genes and 8 ribosomal RNA genes. Most of the protein-coding genes contained a single exon; only 10 genes contained two exons and 4 gene had three exons (i.e., ycf3). The overall GC content of the chloroplast genome was 37.29%, while the corresponding values for the LSC, SSC, and IR regions were 35.33%, 32.39% and 41.03% respectively. Phylogenetic analysis showed that R. nobile clustered with the other three Rheum species included in the study, and Rheum is closely related to Oxyria.
Rheum nobile Hook. f. and Thomson (Polygonaceae) is a giant herbaceous plant, endemic to the alpine zones of the eastern Himalayas at altitudes of between 4000 and 4800 m (Omori and Ohba Citation1996). The most outstanding attribute of this species is its large and showy translucent bracts, which cover the towering inflorescences that measure up to 150 cm in height and are visible from afar. The translucent bracts account for ca. 20% of aboveground biomass during flowering, but they have no capacity for photosynthesis because the chloroplasts they contain have degenerated (Omori et al. Citation2000). R. nobile has been chosen as a model species for investigating alpine adaptation in ‘glasshouse’ plants (Zhang et al. Citation2010). In this study, we sequenced the complete chloroplast (cp) genome of R. nobile (GenBank: MN880789) to provide essential genomic information, and the phylogenetic relationships of the species with other taxa were analyzed.
Fresh leaves of R. nobile were collected from the Qinghai-Tibetan Plateau (29°36′N, 94°39′E; China; voucher LZU-2019-Tibet-24 deposited in the herbarium of Lanzhou University, Lanzhou, China). Genomic DNA was isolated using the modified CTAB method (Doyle Citation1987). Whole-genome sequencing was conducted with 150 bp paired-end reads on the Illumina Hiseq 2500 Platform (Illumina, San Diego, CA). After quality trimming, a total of 217 Gb clean reads were assembled using Fast-Plast pipeline (https://github.com/mrmckain/Fast-Plast). Annotation was performed by Plann (Plastome Annotator), which is suitable for plastome annotation based on a well-annotated sequenced relative (Huang and Cronk Citation2015). We then corrected the annotation and generated the R. nobile plastid genome map with Geneious (Kearse et al. Citation2012). We also constructed phylogenetic trees using RaxML version 8 (Stamatakis Citation2014) with the alignments created by MAFFT (Katoh and Standley, Citation2013) from the plastid genomes of 14 species.
The complete chloroplast genome of R. nobile was a quadripartite circle 161,250 bp in length, comprising a large single copy (LSC) region of 86,290 bp and a small single copy (SSC) region of 12,838 bp, separated by two inverted repeat (IR) regions, each of 31,061 bp (). It contained 127 genes, including 82 protein-coding, 8 ribosomal RNA, and 35 tRNA genes. The plastome contained 93 unique genes, 17 genes duplicated in the IR regions. Among annotated genes, 10 genes contained a single intron, and 4 gene had two introns (i.e., ycf3). The base composition of the chloroplast genome was uneven (31.21% A, 18.98% C, 18.31% G, 31.50% T), with an overall GC content of 37.29%, and the corresponding values for the LSC, SSC and IR regions were 35.33%, 32.39% and 41.03% respectively.
Figure 1. Phylogenetic tree based on 14 complete chloroplast genome sequences. Bootstrap values are shown at each node. The GenBank accession number for each species is given after its name.
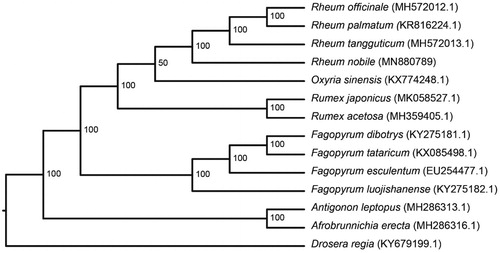
Phylogenetic analysis was carried out using the complete cp genomes of R. nobile and 13 other species, including 12 members of the Polygonaceae and one species from the Droseraceae as outgroup (). The results showed that the four Rheum species clustered together and that Rheum and Oxyria are closely related. This complete chloroplast genome for R. nobile will be of value in gaining an increased understanding of plant adaptation to alpine zones.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley D M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010. 23329690
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Omori Y, Ohba H. 1996. Pollen development of Rheum nobile Hook. f. and Thomson (Polygonaceae), with reference to its sterility induced by bract removal. Bot J Linn Soc. 122:269–278.
- Omori Y, Takayama H, Ohba H. 2000. Selective light transmittance of translucent bracts in the Himalayan giant glasshouse plant Rheum nobile Hook. f. and Thomson (Polygonaceae). Bot J Linn Soc. 132(1):19–27.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang D, Liu B, Zhao C, Lu X, Wan D, Fei M, Litong C, Jianquan L. 2010. Ecological functions and differentially expressed transcripts of translucent bracts in an alpine ‘glasshouse’ plant Rheum nobile (Polygonaceae). Planta. 231(6):1505–1511.
