Abstract
Roscoea alpina is a medicinal plant commonly used in southwest of China. In this study, we sequenced the complete chloroplast (cp) genome sequence of R. alpina to investigate its phylogenetic relationship in the family Zingiberaceae. The chloroplast genome of R. alpina was 163,395 bp in length with 58.9% overall GC content, including a large single copy (LSC) region of 87,739 bp, a small single copy (SSC) region of 16,086 bp and a pair of inverted repeats (IRs) of 29,785 bp. The cp genome contained 113 genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis indicated Roscoea was closely related to Curcuma.
Roscoea is a genus of the Zingiberaceae family, which includes 20 species in the world. Most of them are widely distributed in Bhutan, China, India, Kashmir, Myanmar, Nepal, Sikkim, and Vietnam (Li and Ian Citation1994). There are 14 species distributed in China, mainly in southwest China (Minghua et al. Citation2008). Plants of this genus have been widely used in traditional Chinese medicine for thousands of years (Jiangsu New Medical College Citation1977). Among these species, Roscoea alpina is widely distributed in southwest China which have been used in ethnic medicine as fracture and trauma bleeding (Minru et al. Citation2016). However, up to now for such medicinal plant, many studies have mainly focused on describing its resources and ecology (Paudel et al. Citation2019), with little involvement in its molecular biology, so that no comprehensive genomic resource is conducted for it. Here, we report the chloroplast genome sequence of R.alpina and find its internal relationships within the family Zingiberaceae.
Fresh and clean leave materials of R. alpina were collected from Jiang Li city, Yunnan, China (N27°0′18.82″, E100°12′5.16″), and the plant materials and a voucher specimen (No. TC20) were deposited at Tourism and Culture College of Yunnan University (Lijiang). Total genomic DNA was extracted using the improved CTAB method (Doyle Citation1987; Yang et al. Citation2014), and sequenced with Illumina Hiseq 2500 (Novogene, Tianjin, China) platform with pair-end (2 × 300 bp) library. The raw data was filtered using Trimmomatic v.0.32 with default settings (Bolger et al. Citation2014). Then paired-end reads of clean data were assembled into circular contigs using GetOrganelle.py (Jin et al. Citation2018) with Zingiber officinale (NC_044775) as reference. Finally, the cpDNA was annotated by the Dual Organellar Genome Annotator (DOGMA; http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004) and tRNAscan-SE (Lowe and Chan Citation2016) with manual adjustment using Geneious v. 7.1.3 (Kearse et al. Citation2012).
The circular genome map was generated with OGDRAW v.1.3.1 (Greiner et al. Citation2019). Then, the annotated chloroplast genome was submitted to the GenBank under the accession number MT012418. The total length of the chloroplast genome was 163,395 bp, with 40.6% overall GC content. With typical quadripartite structure, a pair of IRs (inverted repeats) of 29,785 bp was separated by a small single copy (SSC) region of 16,086 bp and a large single copy (LSC) region of 87,739 bp. The cp genome contained 113 genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among the annotated genes, 18 genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, ycf3) contained one intron, whereas another two genes(ycf3 and clpP) possessed two introns.
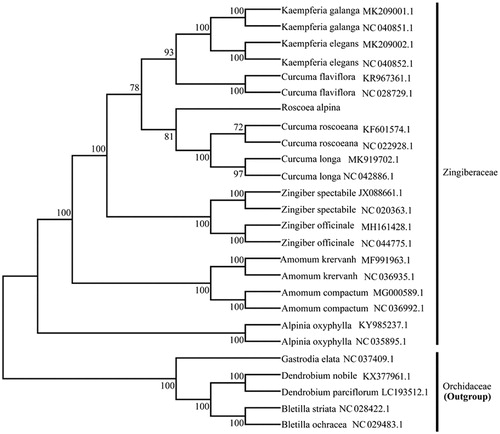
To investigate its taxonomic status, a total of 21 cp genome sequences of Zingiberaceae species were downloaded from the NCBI database used for phylogenetic analysis. After using MAFFT V.7.149 for aligning (Katoh and Standley Citation2013), a neighbor-joining (NJ) tree was constructed in MEGA v.7.0.26 (Kumar et al. Citation2016) with 1000 bootstrap replicates and five Orchidaceae species (Bletilla ochracea NC_029483.1, Bletilla striata NC_028422.1, Dendrobium nobile KX377961.1, Dendrobium parciflorum LC193512.1 and Gastrodia elata NC_037409.1) were used as outgroups. The results showed that Roscoea was closely related to Curcuma (). Meanwhile, the phylogenetic relationship in Zingiberaceae was consistent with previous studies and this will be useful data for developing markers for further studies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Doyle J, 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1. 11–15.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Minru J, Yi Z. 2016. Dictionary of Chinese ethnic medicine. Beijing: China Medical Science and Technology Press.
- Jiangsu New Medical College. 1977. Dictionary of Chinese Materia Medical (In Chinese). Shanghai: Science and Technology Press of Shanghai.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. :1–11.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li HW, Ian CH. 1994. Zingiberaceae. In: Wu ZY and Raven PH, editors. Flora of China (17th). Beijing: Science Press.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Minghua L, Huailong W, Honghui L. 2008. Species and distribution of roscoea in china and their medicinal uses. Chinese Wild Plant Resources. :35–37 + 41.
- Paudel BR, Dyer AG, Garcia JE, Shrestha M. 2019. The effect of elevational gradient on alpine gingers (Roscoea alpina and R. purpurea) in the Himalayas. PeerJ 7:e7503. doi:10.7717/peerj.7503
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.