Abstract
The complete mitochondrial genome (mitogenome) of Zemeros flegyas (Lepidoptera: Riodinidae) is 15,219 bp in length, harboring typical 37 mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and an AT-rich region. The Z. flegyas mitogenome includes a cox1 gene with an atypical CGA(R) start codon and three genes (cox1, nad5, nad4) exhibiting incomplete stop codons. The putative AT-rich region is 385 bp long with several structures characteristic of lepidopteran. Phylogenetic analysis showed that Z. flegyas is closely related to other riodinids. The complete mitogenome data provided here would be useful for further understanding the higher systematics of Riodinidae.
The family Riodinidae shows a remarkable diversity of morphological, ecological and behavioral traits (DeVries Citation1997). Moreover, the phylogenetic position and higher systematics of riodinids remain unresolved and need to be elucidated (Zhao et al. Citation2013; Shen et al. Citation2015; Seraphim et al. Citation2018). Mitochondrial genome (mitogenome) has been widely used as an informative molecular marker for diverse evolutionary study areas (Boore Citation1999; Salvato et al. Citation2008). Within Riodinidae, only two complete mitogenomes have been reported recently (Zhao et al. Citation2013; Kim and Kim Citation2014).
Here, we determined the complete mitogenome of a riodinid, Zemeros flegyas, which was collected from Sanming in Fujian Province, China (coordinates: E117°62′, N26°27′), and kept in the laboratory at −20 °C under the accession number SQH-20160720. Total genomic DNA was extracted from thorax muscle of an adult individual using the Rapid Animal Genomic DNA Isolation Kit (Sangon, China). The raw sequences were assembled and annotated using the BioEdit 7.0 (Hall Citation1999) and MEGA7.0 software (Kumar et al. Citation2016) with reference to the mitogenome of Abisara fylloides (GenBank accession HQ259069).
The complete mitogenome of Z. flegyas is 15,219 bp in length (GenBank accession is MK521434), harboring 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs) and 1 AT-rich region. Its gene arrangement and orientation are identical to those found typically in lepidopteran insects including other riodinids (Wan et al. Citation2013; Zhao et al. Citation2013; Kim and Kim Citation2014). The nucleotide composition is also biased toward AT nucleotides (81.8%). Besides the AT-rich region, 13 intergenic spacers (134 bp in total) and 14 overlapping regions (68 bp in total) are dispersed throughout the whole genome.
All PCGs are initiated by typical ATN, except for cox1 gene, which started with the unusual CGA(R) as observed in most of the other sequenced butterflies (Zhao et al. Citation2013; Kim and Kim Citation2014; Shi et al. Citation2019). Ten PCGs have a complete stop codon, while the remaining three genes (cox1, nad5, nad4) end with a single T. All tRNAs harbor the typical cloverleaf structures commonly found in insects, except for trnS1(AGN), whose dihydrouridine (DHU) arm is replaced by a simple loop. The rrnL is 1326 bp (84.7% AT) and the rrnS is 767 bp (85.6% AT). The 385 bp long AT-rich region exhibits the highest AT content (94.5%) of any region of the whole genome, and contains several structures characteristic of lepidopterans, such as the ATAGA motif followed by a 19-bp poly-T stretch, a microsatellite-like (TA)6 element preceded by the ATTTA motif (Salvato et al. Citation2008; Kim et al. Citation2014).
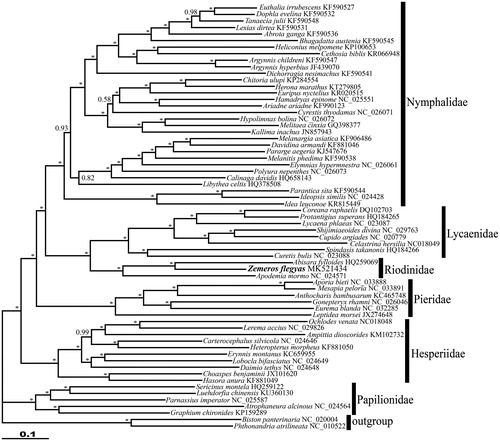
The phylogenetic tree was reconstructed using MrBayes v3.1.2 (Ronquist and Huelsenbeck Citation2003) based on concatenated nucleotide sequences of 13 PCGs and 2 rRNAs (see for details). The phylogenetic analysis revealed that Z. flegyas formed a monophyletic group with other riodinids and yielded the familial relationships (Papilionidae+ (Hesperiidae + (Pieridae + ((Lycaenidae + Riodinidae) + Nymphalidae)))), confirming the recent studies (Kim et al. Citation2014; Shen et al. Citation2015; Espeland et al. Citation2018).
Figure 1. The Bayesian inference (BI) phylogenetic tree of Zemeros flegyas and other butterflies. Phylogenetic reconstruction was done from a concatenated matrix of 13 protein-coding mitochondrial genes and 2 ribosomal RNA genes regions in the mitochondrial genome. The numbers beside the nodes correspond to the posterior probability values (* = 1.00). Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- DeVries PJ. 1997. Butterflies of Costa Rica and their natural history, Volume II: Riodinidae. New Jersey: Princeton University Press.
- Espeland M, Breinholt J, Willmott KR, Warren AD, Vila R, Toussaint EFA, Maunsell SC, Aduse-Poku K, Talavera G, Eastwood R, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Kim MJ, Kim I. 2014. Complete mitochondrial genome of the Mormon metalmark butterfly, Apodemia mormo (Lepidoptera: Riodinidae). Mitochondrial DNA Part A. 27(2):1–3.
- Kim MJ, Wang AR, Park JS, Kim I. 2014. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 549(1):97–112.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genom. 9(1):331.
- Seraphim N, Kaminski LA, Devries PJ, Penz C, Callaghan C, Wahlberg N, Silva-Brandão KL, Freitas A. 2018. Molecular phylogeny and higher systematics of the metalmark butterflies (Lepidoptera: Riodinidae). Syst Entomol. 43(2):407–425.
- Shen JH, Cong Q, Grishin NV. 2015. The complete mitochondrial genome of Papilio glaucus and its phylogenetic implications. Meta Gene. 5:68–83.
- Shi QH, Lin XQ, Ye X, Xing JH, Dong GW. 2019. Characterization of the complete mitochondrial genome of Minois dryas (Lepidoptera: Nymphalidae: Satyrinae) with phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1447–1449.
- Wan XL, Kim MJ, Kim I. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40(11):6333–6349.
- Zhao F, Huang DY, Sun XY, Shi QH, Hao JS, Zhang LL, Yang Q. 2013. The first mitochondrial genome for the butterfly family Riodinidae (Abisara fylloides) and its systematic implications. Zool Res. 34(5):109–119.
