Abstract
We here report the complete chloroplast genome of Pedicularis verticillata and the reconstructed chloroplast genome of Pedicularis cheilanthifolia, the first such genome reported for this genus. The P. verticillata genome is 142,733 bp, including a 24,984-bp inverted repeat (IR) region, 89,723-bp large single-copy (LSC) region, and 12,042-bp small single copy (SSC) region, and the P. cheilanthifolia genome is 147,807 bp (IRs: 24,916 bp; LSC: 82,718 bp; SSC: 15,257 bp). The chloroplast genome of P. verticillata contains 116 genes, including 71 coding genes, 8 rRNAs, and 37 tRNAs. In the genomes of both species, 10 ndh genes are lost and ndhB was identified in the IR region. Additionally, rps16 is also lost in P. verticillata but not in P. cheilanthifolia. The maximum-likelihood phylogenetic tree showed that Pedicularis is a monophyletic and sister group to other hemi-parasitic species in Orobanchaceae.
Pedicularis L. is a core member of Orobanchaceae. The first complete chloroplast genome sequence of Pedicularis genus was reported for P. cheilanthifolia Schrenk in 2017 (Zhang et al. Citation2017). The chloroplast genomes reported for four other Pedicularis species demonstrated drastic changes in genome structure, mainly characterized by the loss of ndh and expansion of inverted repeat (IR) regions (Cho et al. Citation2018; Wu et al. Citation2019). This change in photosynthesis-related genes has emerged as critical for determining the phylogenetic boundary of Pedicularis species. However, resources for understanding their phylogenomic relationships remain limited.
Here, we present the complete chloroplast genome of P. verticillata L., and reconstructed the chloroplast genome of P. cheilanthifolia. Materials of P. verticillata (Voucher specimen: Cho. 1108002, N41°59′52″, E128°01′31″) and P. cheilanthifolia (Voucher specimen: Cho. 1806010, N42°32′3″, E78° 54′41″) were collected from Mt. Baekdu, China, and Mt. Khrebet Kuylyutau, Kyrgyzstan, respectively. The voucher specimens are stored at the BEC herbarium of Chonnam National University. The DNA library was constructed using Illumina TruSeq nano DNA library prep kit and sequenced using the Illumina Miseq platform (LAS, Seoul, Korea). After trimming, clean data were aligned to the P. hallaisanensis genome (NC.037433.1) using Geneious 10.2.3 (Kearse et al. Citation2012). Annotation was performed using DOGMA (Wyman et al. Citation2004), and manually corrected for start and stop codons and for intron/exon boundaries.
The complete, circular chloroplast genome of P. verticillata (GenBank accession no. MT040753) is 142,733 bp long, comprising two IR regions of 24,984 bp separated by a large single-copy (LSC) region (80,723 bp) and small single-copy (SSC) region (12,042 bp). The genome contains 116 genes: 71 protein-coding, eight rRNA and 37 tRNA genes. Almost all ndh genes are pseudogenized, truncated, or deleted, except ndhB of the IR region. The previously reported P. cheilanthifolia genome was only pseudogenized for ndhF (Zhang et al. Citation2017), in contrast to other Pedicularis (Cho et al. Citation2018; Wu et al. Citation2019). Accordingly, we re-identified the P. cheilanthifolia gene content. The chloroplast genome of P. cheilanthifolia (GenBank accession no. MT040754) was 147,807 bp long, which is shorter than reported previously (155,159 bp). The genome comprises two IR regions of 24,916 bp separated by an LSC region (82,718 bp) and SSC region (15,257 bp), with a total of 117 genes: 72 coding genes, 8 rRNAs, and 37 tRNAs. Almost all ndh genes, except for ndhB, were lost, which contrasts with the previous result (Zhang et al. Citation2017). Additionally, loss of rps16 was found in the chloroplast genome of P. verticillata but not P. cheilanthifolia.
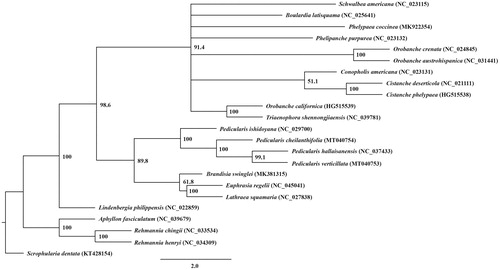
The maximum-likelihood phylogenetic tree was constructed with RAxML v.8.0 (Stamatakis Citation2014) under default parameters and 1000 bootstrap replicates from alignments created by MAFFT (Katoh and Toh Citation2010) using ycf2 sequences (∼6000 bp) from 23 species in Orobanchaceae and Lamiales as the outgroup. In this tree, Pedicularis is a monophyletic and sister group to other hemi-parasitic species in Orobanchaceae (). Rehmannia and Aphyllon (traditionally treated as Scrophulariaceae) are in a sister group to the Orobanchaceae, including Lindenbergia, Orobanche, and Pedicularis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cho WB, Lee DH, Choi IS, Lee JH. 2018. The complete chloroplast genome of hemi-parasitic Pedicularis hallaisanensis (Orobanchaceae). Mitochondrial DNA B. 3(1):235–236.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu C, Fang D, Wei J, Wang X, Chen X. 2019. The complete chloroplast genome of Pedicularis alaschanica (Orobanchaceae). Mitochondrial DNA B. 4(2):2197–2198.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang R, Wang J, Han K, Ren T, Zeng S, Biffin E, Liu ZL. 2017. Complete chloroplast genome sequence of Pedicularis cheilanthifolia, an alpine plant in China. Conserv Genet Resour. 9(4):619–621.