Abstract
The Arctic skate Amblyraja hyperborea is common in the cold water of the Atlantic and Pacific Arctic. Its distribution and taxonomy have been open to discussion. Here, we report on the whole mitochondrial genome sequence of A. hyperborea and the subsequent characterization. The mitogenome is 16,729 bp in length and consists of 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The phylogenetic tree we reconstruct with the mitogenomes of related skates within the order Rajiformes supports the monophyletic assemblage congruent with recent studies, whilst raising the necessity of further investigations to elucidate the relationship among some rajid genera.
Skates belong to the families under the class Chondrichthyes which are well known to have a flattened rhombic body, a thin tail, and enlarged pectoral fins, as well as a cartilaginous skeleton. Among more than 287 skate species distributed worldwide (Ebert and Winton Citation2010), Amblyraja hyperborea is a mesobenthic and bathybenthic species mostly found in the sea floor of the Atlantic and Pacific Arctic (Mecklenburg et al. Citation2016). Given that its observations in the eastern North Pacific were recently documented (Kuhnz et al. Citation2019), A. hyperborea may have a wider distribution than estimated thus far (Mecklenburg et al. Citation2018). The taxonomy of rajid skates has been debatable (Chiquillo et al. Citation2014). The recent molecular phylogenetic analyses have questioned the long-held morphology-based delineation of their taxonomic status. Herein, we sequence and characterize the complete mitogenome of an A. hyperborea specimen bycaught from the Antarctic Ocean. We expect these results will help clarify the taxonomy of the Rajiformes and serve as one of the instrumental resources for the future conservation endeavor.
A bycatch specimen of A. hyperborea was collected from the Antarctic Ocean in 2015 (72°59′00″S, 177°00′00″E) and deposited in the National Marine Biodiversity Institute of Korea (MABIK Lot No. 0000098). Its genomic DNA was isolated following Asahida et al. (Citation1996), and the mitogenome amplified through two independent and overlapping PCR runs was completely sequenced using 25 primers.
The assembled A. hyperborea mitogenome (GenBank MN883843) is a 16,729 bp long circular DNA with 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The gene content and gene order are highly similar to that of the congeneric A. radiata (Rasmussen and Arnason Citation1999), in addition to those of typical vertebrates.
The A. hyperborea mitogenome was aligned to the other species belonging to the Rajiformes whose mitogenomes were acquired from GenBank (http://www.ncbi.nlm.nih.gov/). They were refined after excluding ambiguously aligned regions. We used RAxML 8.2 (Stamatakis Citation2014) to reconstruct a maximum likelihood (ML) tree based on the nucleotide matrix of 12 protein-coding genes excluding nad6, 2 rRNA genes, and 22 tRNA genes. The matrix was broken down into five partitions: 3623, 3623, 3623, 2519, and 1495 bp each for the bases in the codons of protein-coding genes and two regions of rRNA and tRNA genes.
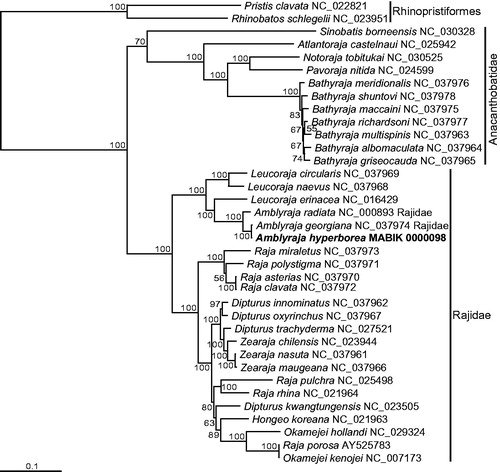
The Rajiformes is monophyletic, which is sufficiently supported by the high bootstrapping value to the outgroup species of the Rhnoprisiformes (). The ingroup is referred to as two explicitly partitioned lineages (the families Anacanthobatidae and Rajidae), each reflecting a monophyly. Amblyraja hyperborea is closely related to congeneric A. georgiana, and these two species along with A. radiata are grouped into a single clade. All of the above are consistent with the comprehensive phylogenetic analysis based on three mitochondrial loci (Chiquillo et al. Citation2014). Interestingly, a few specimens within the Rajidae such as Dipturus, Raja, and Okamejei are associated with different taxa from the ones in the previous study. Our sampling site also backs up the existence of A. hyperborea in the south of the Equator.
Figure 1. Mitogenome-based maximum likelihood (ML) phylogram of Amblyraja hyperborea (in bold), plus related species. The bars on the right represent the outgroup order (Rhinopristiformes) and the two families (Anacanthobatidae and Rajidae) within the order Rajiformes. The nucleotide sequence matrix included the codons of the 12 protein-coding, rRNA, and tRNA genes. A bootstrap value above 50% in the ML analysis is indicated at each node.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish Sci. 62(5):727–730.
- Chiquillo KO, Ebert DA, Slager CJ, Crow KD. 2014. The secret of the mermaid’s purse: Phylogenetic affinities within the Rajidae and the evolution of a novel reproductive strategy in skates. Mol Phylogenet Evol. 75:245–251.
- Ebert DA, Winton MV. 2010. Chondrichthyans of high latitude seas. In: Carrier JC, Musick JA, Heithaus MJ, editors. Sharks and their relatives II – biodiversity, adaptive physiology and conservation. Boca Raton: CRC Press; p. 116–158.
- Kuhnz LA, Bizzarro JJ, Ebert DA. 2019. In situ observations of deep-living skates in the eastern North Pacific. Deep-Sea Res Pt I. 152:103104.
- Mecklenburg CW, Mecklenburg TA, Sheiko BA, Steinke D. 2016. Pacific Arctic marine fishes. Akureyri, Iceland: Conservation of Arctic Flora and Fauna.
- Mecklenburg CW, Lynghammar A, Johannesen E, Byrkjedal I, Christiansen JS, Dolgov AV, Karamushko OV, Møller PR, Steinke D, Wienerroither RM. 2018. Marine fishes of the Arctic region. Akureyri, Iceland: Conservation of Arctic Flora and Fauna.
- Rasmussen AS, Arnason U. 1999. Molecular studies suggest that cartilaginous fishes have a terminal position in the piscine tree. Proc Natl Acad Sci USA. 96(5):2177–2182.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
