Abstract
The silver-studded blue butterfly Plebejus argus (Lepidoptera: Lycaenidae) has a mitochondrial genome of 15,426 bp. It consists 13 protein-coding genes, 23 tRNAs with an extra copy of tRNASer, two ribosomal RNA genes, and an AT-rich control region. Using whole mitogenome alignments, we reconstructed the phylogenetic relationships of 23 butterfly species within six families (11 taxa are from Lycaenidae). The maximum-likelihood (ML) tree topology was consistent with previous studies, which suggested a close relationship between the genus Plebejus and Cupido.
Lycaenidae, also called gossamer-winged butterflies, is the second-largest butterfly family including ∼6000 species worldwide (Pierce et al. Citation2002). The majority of lycaenid species are mutually associated with ants (Pierce et al. Citation2002; Espeland et al. Citation2018), although understanding the evolutionary pattern of such interactions depends upon the uncover of robust butterfly relationships. Recently, the accumulation of whole mitochondrial genomes has facilitated phylogenetic studies (Burger et al. Citation2003; Cameron Citation2014; Liang et al. Citation2018). Here, we presented the first complete mitogenome of Plebejus argus (Lepidoptera: Lycaenidae), which is widely distributed across the Palearctic. The phylogenetic position of P. argus was also estimated using published whole mitogenomic data.
An individual of P. argus was collected from Tieqiao Forest Farm (E113.34, N37.45), Heshun County, Shanxi Province, China in 9 May 2019 (voucher number: BL_YZ_DHD_002, kept in Nanjing Forestry University). Following Chen et al. (Citation2020), total genomic DNA was extracted and sequenced on an Illumina HiSeq X Ten platform. The whole mitogenome of P. argus was assembled in Geneious Prime® 2019.2.1 through mapping filtered reads to reference mitogenomes (Coreana raphaelis, GenBank: NC_007976 and Celastrina hersilia, GenBank: NC_018049), with ∼1400× coverage. We then performed automatic annotation using the MITOS web server (Bernt et al. Citation2013), and manually verified the gene boundaries by comparing sequence annotations among closely related species.
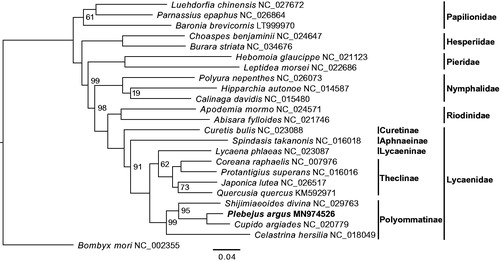
To explore the phylogenetic position of P. argus, we collected mitogenomes of ten additional lycaenid species and 13 outgroups from Genbank. After excluding the extremely gappy regions (i.e. column occupancy < 0.5) from the whole mitogenomic alignments, a maximum likelihood (ML) tree was built in RAxML v8.2.10 (Stamatakis Citation2014) with the GTRGAMMA model and 200 ultrafast bootstraps (-f a). The ML tree highly supported (100%) a sister relationship between P. argus and Cupido argiades (), and a monophyletic clade of all lycaenid butterflies that further group with Riodinidae. The overall topology is also consistent with previous trees based on hybrid capture-based genomic data (Espeland et al. Citation2018), morphological and small scale molecular data (Heikkilä et al. Citation2012).
Figure 1. Inferred phylogenetic relationships among Lycaenidae and allies using mitogenomic data. The domestic silkworm Bombyx mori was used as the outgroup. Bootstrap values at major nodes are 100% unless indicated on the tree. GenBank accession numbers of all species used in this study are shown by the species name.
The complete mitochondrial genome of P. argus is circular and 15,426 bp in length with 17.9% GC content. It encodes 38 genes including 13 protein-coding genes, the standard 22 tRNAs, an extra trnS1-AGT tRNA (adjacent to the original trnS1-AGA tRNA), two ribosomal RNA genes, and a putative control region (GenBank accession number: MN974526). The duplication of mitochondrial tRNAs is a rare genomic change (RGC), although it has been identified in a wide array of animal classes (Kim et al. Citation2012; Kumazawa et al. Citation2014; Oceguera-Figueroa et al. Citation2016). RGCs including duplication and remolding of mitochondrial sequences might provide a valuable tool for systematic studies (Rokas and Holland Citation2000; San Mauro et al. Citation2006).
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 69(2):313–319.
- Burger G, Gray MW, Lang BF. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19(12):709–716.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Chen G, Wu H, Wang N, Zhong S, Zhou Y, Liang B. 2020. A mitogenomic phylogeny of spiders and complete mitochondrial genome of Cyriopagopus hainanus (Araneae: Theraphosidae). Mitochondrial DNA Part B. 5(1):782–783.
- Espeland M, Breinholt J, Willmott KR, Warren AD, Vila R, Toussaint EFA, Maunsell SC, Aduse-Poku K, Talavera G, Eastwood R, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.
- Heikkilä M, Kaila L, Mutanen M, Pena C, Wahlberg N. 2012. Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc R Soc B. 279(1731):1093–1099.
- Kim S, Kim J, Choi HG, Park JK, Min GS. 2012. Complete mitochondrial genome of the northern mauxia shrimp Acetes chinensis (Decapoda. Dendrobranchiata, Sergestoidae). Mitochondrial DNA. 23(1):28–30.
- Kumazawa Y, Miura S, Yamada C, Hashiguchi Y. 2014. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genomics. 15(1):930.
- Liang B, Wang N, Li N, Kimball RT, Braun EL. 2018. Comparative genomics reveals a burst of homoplasy-free numt insertions. Mol Biol E. 35(8):2060–2064.
- Oceguera-Figueroa A, Manzano-Marin A, Kvist S, Moya A, Siddall ME, Latorre A. 2016. Comparative mitogenomics of leeches (Annelida: Clitellata): genome conservation and Placobdella-specific trnD gene duplication. PloS One. 11(5):e0155441.
- Pierce NE, Braby MF, Heath A, Lohman DJ, Mathew J, Rand DB, Travassos MA. 2002. The ecology and evolution of ant association in the Lycaenidae (Lepidoptera). Annu Rev Entomol. 47(1):733–771.
- Rokas A, Holland PW. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol E. 15(11):454–459.
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. 2006. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol E. 23(1):227–234.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
