Abstract
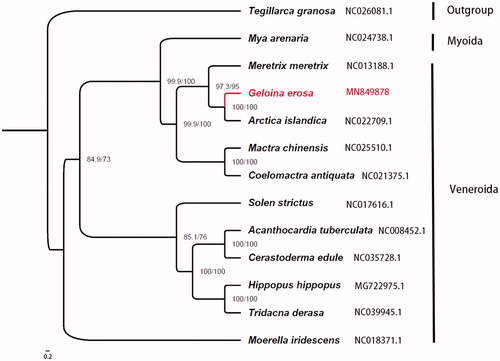
We assembled one mitochondrial genome of Geloina erosa from NovaSeq 6000 sequencing data. The complete mitochondrial genome of G. erosa was 15,041 bp in length, including 36 genes, which are 12 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes ((12S and 16S), which showed the typical marine bivalves mitochondrial gene composition. The overall base composition was 21.16% for A, 46.11% for T, 7.73% for C, and 25% for G. A phylogeny of 13 bivalves showed G. erosa was clustered within Veneroida.
Keywords:
Mangrove wetlands represent a special ecosystem that spans across land and ocean (Feller et al. Citation2010). Shellfishes are an important component of the benthos in mangrove, and in the mangrove of southern China, Geloina erosa is a dominant species which has highly abundant biomass (Zhou Haolang et al. Citation2014). Geloina erosa is a member of Corbiculidae, and to date, little mitochondrial and ribosomal sequences have been reported for Corbiculidae (NCBI, accessed 2020 Jan 22). To be helpful for assessing its systematic position, here we assembled the complete mitochondrial genome of Geloina erosa, which is the first mitogenome sequence in Corbiculidae.
In this study, samples were obtained from mangrove wetland in Beihai, Guangxi, China (21.57°N, −109.16°E) and the specimens were vouchered at GMRC (specimen Accession number GE#60-70). The voucher specimen and extracted DNA were stored at −20 °C in the Guangxi Mangrove Research Center, China. Total genomic DNA was obtained from muscle of individual using the QIAamp DNA Micro Kit (Qiagen, Germany) and sequenced by NovaSeq 6000. The mitogenome was assembled with NOVOPlasty v2.7.0 (Dierckxsens et al. Citation2017), annotated with MitoZ v2.4 (Meng et al. Citation2019), and deposited in the GenBank with an accession number MN849878.
The complete mitochondrial genome of G. erosa was 15,041 bp in length, including 36 genes, which are 12 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes ((12S and 16S), and all of them were located on the positive strand. Although the typical ATP8 gene was not found in this study, a truncated ATP8 which was located between 7039 and 7156 codons, contained an ORF encoding for only 39 amino acids. Similarly, truncated ATP8 genes were also reported in the mitochondrial DNA genomes of other marine bivalves, like Paphia euglypta (Xu et al. Citation2010) and Venerupis philippinarum (Dreyer and Steiner Citation2006). The base composition was 21.16% for A, 46.11% for T, 7.73% for C, and 25% for G, which showed a bias of higher A + T content (67.27%) than the G + C content (32.73%). For the 12 protein-coding genes, seven of them used TAA as the stop codon, one (ND1) used TAG, and the other three (COX1, COX2 and ND4L) used uncompleted T or TA.
Amino acid sequences were aligned using MAFFT v7.407 (Katoh and Standley Citation2013) and trimmed with trimAl v1.4.1 (Capella-Gutierrez et al. Citation2009) with the heuristic method ‘automated1’. The phylogenetic tree of 13 sequences including G. erosa, the other 11 species from Veneroida and Myoida, and one Arcoida specie as an outgroup was reconstructed using IQTREE v1.6.10 (Nguyen et al. Citation2015) with the partitioning method ().
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dreyer H, Steiner G. 2006. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica – and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 3(1):13.
- Feller IC, Lovelock CE, Berger U, McKee KL, Joye SB, Ball MC. 2010. Biocomplexity in mangrove ecosystems. Annu Rev Mar Sci. 2(1):395–417.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(1):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Xu X, Wu X, Yu Z. 2010. The mitogenome of Paphia euglypta (Bivalvia: Veneridae) and comparative mitogenomic analyses of three venerids. Genome. 53(12):1041–1052.
- Zhou Haolang ZJ, Xing Yongze WB, Bin Y. 2014. Characteristics of distribution and the influential factors of Mangrove Clam, Polymesoda erosa (Solander 1786), in Guangxi. Guangxi Sci. 21(2):147–152.