Abstract
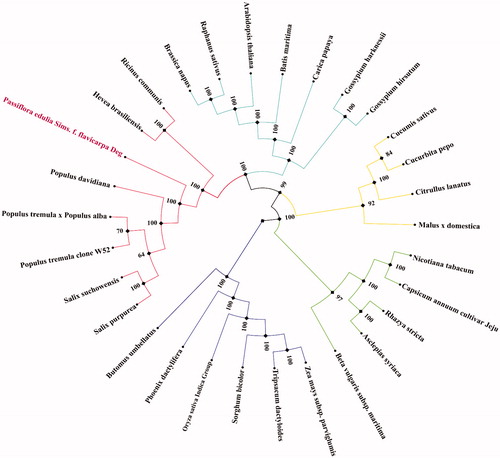
The passion fruit (Passiflora edulis Sims), also called as ‘Baixiang-guo’, has been widely studied and grown, becoming the new darling of the fruit market throughout southern China. In the present study, we found the complete mitochondrial genome of the yellow passion (Passiflora edulis Sims. f. flavicarpa Deg.) fruit to be 680,480 bp in length. A total of 74 genes were predicted, including 41 protein-coding genes, 3 ribosomal RNA genes, and 30 transfer RNA genes. Under phylogenetic analysis, yellow passion fruit clustered with Populus davidiana, which further confirmed the traditional classification.
The passion fruit, genus Passiflora, belongs to the family Passifloraceae, which has more than 500 species (Rodríguez and Margarita Citation2004). The yellow passion fruit is the best known of the edible and marketable species (Salazar et al. Citation2016). It is mainly native to South America, principally in Brazil, which is the largest passion fruit producer in the world. The yellow passion fruit is one of the most economically important species of passion fruits in Brazil, accounting for 95% of the national production (Gioppato et al. Citation2018; Gomes et al. Citation2019). In recent decades, the yellow passion fruit has gained great appreciation from the people in China due to its unique taste and high level of antioxidants. To further elucidate the evolution and diversity of species in the genus Passiflora and family Passifloraceae, we here report the complete mitochondrial genome of the yellow passion fruit.
Fresh, young samples of the yellow passion fruit were collected from the Fruit Research Institute, Fujian Academy of Agricultural Sciences (Fujian, China, 119°19′57″E, 26°7′47″N), in Fuzhou, China, and deposited at Fujian Agriculture and Forestry University (No.FAFUYSJ01). We extracted the total genomic DNA using the optimized CTAB protocol and built an Illumina paired-end library with 500 bp insert size. The library was sequenced using an Illumina HiSeq 2000 platform system (Illumina, San Diego, CA, USA) at Beijing Genomics Institute (BGI, Shenzhen, China), and it generated ∼4.51 G of raw data. After filtered by the FastQC software (Andrews Citation2014), high-quality clean reads of ∼4.5 G were employed for de novo assembly using SPAdes v 3.9.0 (http://bioinf.spbau.ru/spades) (Bankevich et al. Citation2012). The cp mitochondrial genome sequence was annotated using the online program GeSeq (Tillich et al. Citation2017).The annotated mitochondrial genome was submitted to GenBank under accession number MT140634.
The total length of the mitochondrial genome was 680,480 bp with 44.65% GC content, and the nucleotide contents of A, T, G, and C were 27.85%, 27.5%, 22.5%, and 22.15%, respectively. A total of 74 genes were predicted, comprising 41 protein-coding genes, 3rRNA genes, and 30 tRNA genes. The protein-coding gene regions were found to account for 64.7% of the complete mitochondrial genome and a total length of 440,290 bp, while tRNA genes and rRNA genes made up 2218 and 5454 bp of the genome, respectively. Among all genes, five genes (nad1, nad2, nad4, nad5, and nad7) had exon-intron structure. The phylogenic maximum-likelihood trees were constructed using RaxML software v 8.2.9, of which the bootstrap values were calculated using 1000 replicates (Stamatakis Citation2014), based on 30 mitochondrial genomes from plant species. These results () revealed Passiflora edulis Sims. f. flavicarpa Deg clustered with Populus davidiana, which is within Salicaceae. This was consistent with the findings of a previous study (Cauz dos Santos et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Andrews S. 2014. FastQC: a quality control tool for high throughput sequence data. [online]. Version 0.11.8. UK: Babraham Bioinformatics. [Updated 2018 Oct 4; accessed 2020 February 10] http://wwwbioinformaticsbabrahamacuk/projects/fastqc/
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cauz dos Santos L, Munhoz C, Rodde N, Cauet S, Santos A, Penha H, Dornelas M, Varani A, Oliveira G, Bergès H, et al. 2017. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 8:334.
- Gioppato H, Silva M, Carrara S, Palermo B, Moraes T, Dornelas M. 2018. Genomic and transcriptomic approaches to understand Passiflora physiology and to contribute to passionfruit breeding. Theor Exp Plant Physiol. 31:173–181.
- Gomes FR, de Souza PHM, Costa MM, Sena-Júnior DGD, Souza ALP, Azevedo VM, Carneiro LC, Cruz SCS, Costa CHMD, Silva DFP. 2019. Quality of the pulp of passion fruit produced in the Brazilian Savanna. JAS. 11(4):470.
- Rodríguez RR, Margarita PD. 2004. Aislamiento y cultivo de protoplastos en maracuyá isolation and cultive of protoplast in passion fruit. Acta Biol Colombiana. 9:35–46.
- Salazar A, Silva D, Bruckner CH. 2016. Effect of two wild rootstocks of genus Passiflora L. on the content of antioxidants and fruit quality of yellow passion fruit. Bragantia. 75(2):164–172.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.