Abstract
Although the traditional Folmer and Prosser primers enable successful amplification of COI ‘barcoding’ sequences in freshwater zooplankton species, many freshwater zooplankters including Phyllodiaptomus tunguidus are difficult to be amplified using traditional primers. Phyllodiaptomus tunguidus is an endemic calanoid copepod species, widely distributed in (sub)tropical China that plays an important role in freshwater ecosystems. In this study, we developed a new pair of primers for P. tunguidus with significantly higher amplification success rate. The new primers will facilitate the recovery of barcode data from this and probably other copepod species, and stimulate further studies of phylogeny and genetic diversity on P. tunguidus.
Introduction
Copepods are abundant and diverse in freshwaters, occurring as a major component of most planktonic, benthic and groundwater metazoan communities (Dussart and Defaye Citation2001, Citation2002). Copepods often dominate zooplankton communities, play a crucial role as primary and secondary consumers in aquatic ecosystems, and are a major food source for other aquatic predators. Calanoid copepods (Copepoda: Calanoida) in inland waters are principally represented by the family Diaptomidae (Huys and Boxshall Citation1991; Boxshall and Jaume Citation2000; Dussart and Defaye Citation2001). Many genera of Diaptomidae are endemic to particular continents or parts of continents. Phyllodiaptomus conforms to this pattern and has its highest species richness in Southeast Asia (Sanoamuang and Teeramaethee Citation2006). Phyllodiaptomus tunguidus (Copepod, Calanoida, Diaptomidae) is endemic to China, and widely distributes in reservoirs and ponds of (sub)tropical China. It is the dominant species of zooplankton in most reservoirs in southern China (Lin et al. Citation2003; Zhao and Han Citation2007). In spite of its importance, there is no published molecular information on this keystone species.
The mitochondrial COI gene fragment known as the barcoding sequence is a widely used DNA marker for species identification in the ongoing ‘barcoding of life’ project and proved to be a suitable marker for species identification in crustaceans (Hajibabaei et al. Citation2005; Costa et al. Citation2007). However, successful amplifications of COI are not always achieved in freshwater zooplankton species. Currently, ‘Folmer’ (Folmer et al. Citation1994) and ‘Prosser’ primers (Prosser et al. Citation2013) are the most frequently used universal primers for Copepods. Oddly, in P. tunguidus the gene appears to be more difficult to amplify than in most other copepods when using those universal primers. Therefore, our aim of this work was to develop a specific pair of primers to amplify the COI barcoding fragment of P. tunguidus reliably. This primer pair will greatly promote further studies in phylogeny and population genetics of P. tunguidus. Possibly, it may apply to other species as well.
Material and methods
Primer design
The complete mitochondrial genome of P. tunguidus was sequenced and uploaded to GenBank database (accession number: MN927223) in 2020, and the COI genes were excised and aligned with COI sequences of Phyllodiaptomus sp. (downloaded from GenBank database, accession number: JN183940) using BioEdit (Hall Citation1999). Potential primer regions were analyzed using Primer version 6.0 (Primer Biosoft Inst., Palo Alto, CA) with default parameters. A new primer set was then manually designed based on flanking the more polymorphic regions of the COI gene.
DNA extraction, PCR and sequencing
Whole genomic DNA was extracted from 24 specimens of P. tunguidus collected from the Qingshitan (25.30∘ N, 110.09∘ E) and Gaozhou reservoirs (22.23∘ N, 111.03∘ E) in southern China, using Chelex 100 (Bio-Rad Laboratories, CA, USA). All collected specimens and extracts were stored at −20 °C at the Institute of Hydrobiology, Jinan University, Guangzhou, China.
Two populations of P. tunguidus were used to test the effectiveness of the new primer pair, Gaozhou population from tropical and Qingshitan population from subtropical China. Of these two populations, DNA extracts from 12 specimens were amplified. Validation of DNA extraction were done with CYTB specific primers (L10319/H10648; Machida et al. Citation2004). In addition, we compared the success rate of the new pair of primers to two other pairs, Folmer (LCO1490/HCO2198) and Prosser (ZplankF1/ZplankR1) (Folmer et al. Citation1994; Prosser et al. Citation2013). PCR amplification was carried out in PCR solution with 3 μl 10 PCR buffer, 1.2 μl dNTPs, 0.5 μl of 20 μM solution of each primer, 21.5 μl ddH2O, 0.3 μl Taq DNA polymerase, 3 μl template. The PCR conditions for amplification were as follows: initial denaturation 1 min at 95 °C, followed by five cycles of 94 °C [for 40 s, 45 °Cfor 40 s, 72 °C for 1 min], then 35 cycles of 94 °C [for 40 s, 51°Cfor 40 s, 72 °C for 1 min] and final extension of 72 °C for 7 min.
PCR products were visualized on a 1.5% agarose gel using Gel DocTM XR + (Bio-Rad, Hercules, CA), positive PCR products were sequenced on an ABI 3730XL automated sequencer with both forward and reverse of Phyllodiaptomus-specific primers.
Results
The new primers are: forward PhylloF (5′-CCAATCGCCAACATAGCATAAA-3′) and reverse PhylloR (5′-AGATATGGCTTTCCCTCGAATAA-3′). They were designed inside the most conservative region of the COI gene, and based on the completed mitochondrial genome of P. tunguidus.
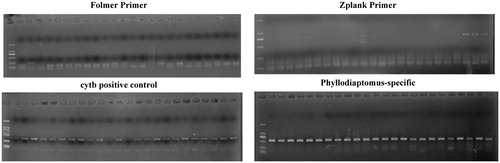
To compare the efficacy of our new primer set to traditional Folmer primers and Zplank primers, we extracted DNA from 24 specimens of two different regions. All of these 24 samples from two populations generated positive PCR products with CYTB primers. The PCR success rate for traditional ZP primers was 29% and no positive band available for Folmer primers. As predicted, the new Phyllodiaptomus-specific primers were highly effective (100%) in amplifying the target region of COI (). Sequences with 573 bp read length and of high quality (). All sequences obtained were uploaded to the GenBank database (accession number: MN912844-MN912862).
Discussion
The COI appears to possess a greater range of phylogenetic signal than other mitochondrial genes (Hebert et al. Citation2003). Compared to other protein-coding genes, its third-position nucleotides show a high number of nucleotide substitutions, and evolved at a higher rate than (about three times greater) that 12S or 16S rDNA (Knowlton and Weigt Citation1998). DNA barcoding provides a unique and effective method for species identification, and is becoming a standard tool in taxonomical and population genetic studies. But, large-scale DNA barcoding studies of freshwater zooplankton are still too rare, mostly due to failures in COI amplification success rates. The success rate can range from 0% to 40%, while some exceptions (Elías-Gutiérrez et al. Citation2008; Jeffery et al. Citation2011). Success rates above 50% have rarely been reported. Compared with Folmer primer, primers for zooplankton can improve PCR success rate for Diaptomus. For population genetics studies, these low PCR success rates still represent a big impediment.
Our new Phyllodiaptomus-based primers improve PCR success rates significantly when compared with traditional Folmer primers. We found that either the traditional primers could only amplify the nonspecific bands, or the target bands were accompanied by many nonspecific bands. Therefore, a combination of primer binding efficiency and primer specificity may explain the performance differences among the Folmer primer, zooplankton primer, and Phyllodiaptomus-specific primer sets. This new primer set will therefore facilitate further studies, at least in calanoid copepods.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boxshall GA, Jaume D. 2000. Making waves: the represented colonization of freshwater by copepod crustaceans. Adv Ecol Res. 31:61–79.
- Costa FO, deWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PD. 2007. Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci. 64(2):272–295.
- Dussart B, Defaye D. 2001. Introduction to Copepods. 2nd edn. Leiden: Backhuys Publishers.
- Dussart B, Defaye D. 2002. World directory of Crustacea Copepoda of inland waters. Leiden: Backhuys Publishers.
- Elías-Gutiérrez M, Martínez-Jerónimo F, Ivanova NV, Valdez-Moreno M. 2008. DNA barcodes for Cladocera and Copepoda from Mexico and Guatemala, highlights and new discoveries. Zootaxa. 1849:1–42.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for a amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 3(5):294–299.
- Hajibabaei M, deWaard JR, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, Mackie PM, Hebert PDN. 2005. Critical factors for assembling a high volume of DNA barcodes. Phil Trans R Soc B. 360(1462):1959–1967.
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41:95–98.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B. 270(1512):313–321.
- Hebert PDN, Ratnasingham S, de Waaard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Roy Soc London B. 270:96–99.
- Huys R, Boxshall GA. 1991. Copepod evolution. London: The Ray Society.
- Jeffery NW, Elías-Gutiérrez M, Adamowicz SJ. 2011. Species diversity and phylogeographical affinities of the Branchiopoda (Crustacea) of Churchill, Manitoba, Canada. PLOS One. 6(5):e18364.
- Knowlton N, Weigt LA. 1998. New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc Lond B. 265(1412):2257–2263.
- Lin QQ, Duan SS, Hu R, Han B-P. 2003. Zooplankton distribution in trophic reservoirs, south China. Internat Rev Hydrobiol. 88(6):602–613.
- Machida RJ, Miya MU, Nishida M, Nishida S. 2004. Large-scale gene rearrangements in the mitochondrial genomes of two calanoid copepods Eucalanus bungii and Neocalanus cristatus (Crustacea), with notes on new versatile primers for the srRNA and COI genes. Gene. 332:71–78.
- Prosser S, Martínez-Arce A, Elías-Gutiérrez M. 2013. A new set of primes for COI amplification from freshwater microcrustaceans. Mol Ecol Resour. 13(6):1151–1155.
- Sanoamuang L, Teeramaethee J. 2006. Phyllodiaptomus thailandicus, a new freshwater copepod (Copepoda, Calanoida, Diaptomidae) from Thailand. Crustaceana. 79(4):475–487.
- Zhao SY, Han B-P. 2007. Structural analysis of zooplankton community in a large deep oligotrophic reservoir – Xinfengjiang Reservoir, South China. J Lake Sci. 19:305–314.