Abstract
Here, we sequenced and annotated the complete mitogenome of Sympetrum striolatum (Odonata: Libellulidae). This mitogenome is 15,435 bp in length, consisting of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA unit genes, and a large non-coding region (putative control region). The S. striolatum mitogenome with an A + T content of 71.54% presented a positive AT-skew (0.081) and a negative GC-skew (−0.127). Twelve PCGs started with a typical ATN codon, the remaining one PCG started with TTG (nad1). All tRNAs had a typical secondary cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm. The Bayesian phylogenetic tree of 29 Odonata species based on the concatenated nucleotide sequences of 13 PCGs supported the monophyly of Libellulidae and a closer relationship of S. striolatum and Brachythemis contaminata.
Keywords:
The Libellulidae is the largest family within Odonata and contains around 1000 species. To better understand the diversity and evolution of Odonata, we determined the complete mitogenome of S. striolatum by using next-generation sequencing through the Illumina NovaSeq platform. Adult specimens were collected from Altay Region (87.55°E, 47.71°N), Xinjiang Uygur Autonomous Region, China, in June 2019. All samples (LZUALT56) have been deposited in the College of Pastoral Agricultural Science and Technology, Lanzhou University, Lanzhou, China. The total genomic DNA was extracted from a single specimen (LZUALT56_1) using a DNeasy Tissue Kit (Qiagen).
The complete mitogenome of S. striolatum was a typical circular DNA molecule with 15,435 bp in length (GenBank accession no. MT075809). This mitogenome contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA unit genes (rrnL and rrnS), and a large non-coding region (putative control region). The order and orientation of the mitochondrial genes was identical to the inferred ancestral arrangement of insects (Boore Citation1999). Gene overlaps were found at nine gene junctions and involved a total of 29 bp, ranging in size from 1 to 8 bp. A total of 59 bp intergenic spacers were present in 11 positions, ranging in size from 1 to 22 bp.
The nucleotide composition of the S. striolatum mitogenome was biased toward A and T, with an A + T content of 71.54%. This mitogenome presented a positive AT-skew (0.081) and a negative GC-skew (−0.127), as found in most insect mitogenomes. The rrnL was 1279 bp long with an A + T content of 75.53%, and the rrnS was 753 bp with an A + T content of 73.31%. Among the 13 PCGs, the lowest A + T content was 64.45% in cox1, while the highest was 77.93% in nad6. Twelve PCGs started with a typical ATN codon: one (nad6) with ATC, one (cox1) with ATA, four (nad2, atp8, nad3, nad5) with ATT, six (cox2, atp6, cox3, nad4, nad4L, cob) with ATG. The remaining PCG started with TTG (nad1). Four PCGs terminated with TAA, one terminated with TA, whereas the remaining eight terminated with an incomplete stop codon T. All of the 22 tRNAs, ranging from 60 bp (trnI) to 72 bp (trnK), had a typical cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm.
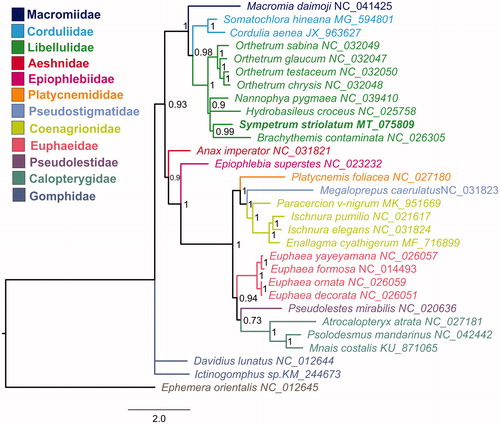
The concatenated nucleotide sequences of 13 PCGs from 29 Odonata species, representing twelve families, were used in phylogenetic analysis. Ephemera orientalis from Ephemeroptera were used as the outgroup. Phylogenetic analysis was performed with PhyloBayes MPI 1.7 b on the CIPRES webserver (Miller et al. Citation2010). The Bayesian phylogenetic tree supported the monophyly of Libellulidae and a closer relationship of S. striolatum with Brachythemis contaminata ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE); 14 November 2010; New Orleans: IEEE.