Abstract
The complete chloroplast (cp) genome of Actinidia valvata (A. valvata) was sequenced and assembled into a circular genome of 156,548 bp in length using Illumina paired-end data. The genome consisted of a pair of inverted repeats (IR) (23,378 bp each) separating two single-copy regions, the large single-copy region (LSC) 88,397 bp and the small single-copy region (SSC) 21,395 bp. The cp genome encoded a total of 131 genes, including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The phylogenetic analysis showed that A. valvata was sister to Actinidia tetramera (A. tetramera).
Actinidia valvata, affiliated to the family Actinidiaceae, is a shrub mainly growing in eastern China (Zhejiang and Jiangxi province) and has a series of applications in traditional Chinese medicine and folk herb (Qu, Xin, et al. Citation2012). The root of A. valvata, commonly known as ‘Mao ren shen’ in traditional Chinese medicine, exhibited remarkable anti-inflammatory and anti-tumoral activities, which has been used for the treatment of lung carcinoma, hepatoma and myeloma for many years (Wang et al. Citation2005; Qu, Zheng, et al. Citation2012). The present study assembled and characterized the complete cp genome of A. valvata, which would provide important genomic resources for promoting its conservation and management.
Plant material of A. valvata was collected from South China Botanical Garden (23°11′0.47″N, 113°21′37″E). The voucher specimen was deposited at the Herbarium of Nanjing Forestry University (accession number: 20160415AV03). Total genomic DNA was extracted from fresh leaves the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and then sequenced using the Illumina Hiseq 2000 platform. The raw sequencing data were filtered and trimmed by fastp program (Chen et al. Citation2018), and then fed into NOVOPlasty version 3.7.2 (Dierckxsens et al. Citation2017) for assembly using the cp genome of A. tetramera (GenBank accession: NC_031187.1) as seed and reference. The assembled genome was then annotated using Plastid Genome Annotator (PGA) (Qu et al. Citation2019) and corrected manually using Macvector v17.0.7. The annotated cp genome of A. valvata was submitted to GenBank with accession number MN602331.
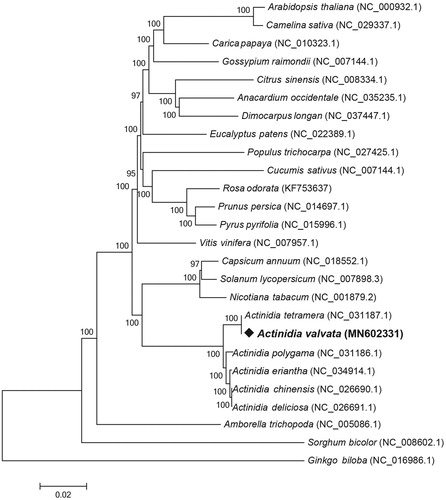
The complete cp genome of A. valvata has a total length of 156,548 bp, consisting of 88,397 bp LSC region and 21,395 bp SSC region separated by a pair of 23,378 bp IR regions. The overall GC content of the whole cp genome is 37.23%, a very common value in family Actinidiaceae. The GC content of the IR regions was 43.2%, which is higher than that of LSC (35.51%) and SSC (31.31%). According the annotation results, the cp genome contains 131 genes, comprising of 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Nineteen genes were found to contain one intron (7 tRNA genes: trnK-UUU, trnL-UAA, trnV-UAC, trnI-GAU × 2, and trnA-UGC × 2; 12 protein-coding genes: rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB × 2, rps12 × 2, and ndhA), and only one gene contains two introns (ycf3). In order to clarify the phylogenetic position of A. valvata, we selected other 25 plant cp genomes with 78 conserved protein-coding genes to reconstruct a Neighbor-Joining (NJ) tree using MEGA X with 1000 bootstrap replicates () (Kumar et al. Citation2018). The reconstructed NJ tree strongly supported that A. valvata was evolutionarily closest to A. tetramera, and then other Actinidia species. These results will benefit us to further study the role of A. valvata in medicine research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Qu L, Xin H, Zheng G, Su Y, Ling C. 2012. Hepatoprotective activity of the total saponins from Actinidia valvata dunn root against carbon tetrachloride-induced liver damage in mice. Evidence-Based Complement Alternat Med. 2012:1–13.
- Qu L-P, Zheng G-Y, Su Y-H, Zhang H-Q, Yang Y-L, Xin H-L, Ling C-Q. 2012. New triterpenoids with cytotoxic activity from Actinidia Valvata. IJMS. 13(12):14865–14870.
- Wang Z, Song Y, Hu J. 2005. Morphological identification and the clinical application of the roots of Actinidia chinensis and Actinidia valvata. Pharm Care Res. 5(2):134.