Abstract
Gentiana manshurica Kitag is a perennial medicinal plant with high economic value. In this study, we first reported the complete chloroplast (cp) genome of G. manshurica. The total length of the cp genome was 149,185 bp in length, consisting of a large single-copy (LSC) region of 81,347 bp, a small single-copy (SSC) region of 17,268 bp, and a pair of inverted repeats (IRs) regions of 25,285 bp each. The cp genome contained 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 37.61%. The phylogenetic analysis indicated that the G. manshurica belongs to the tribe Gentianeae, and showed a close relationship with other Gentiana species.
Gentiana manshurica Kitag belongs to the genus of Gentiana in the family Gentianaceae, which is widely distributed in northeastern China and is a reputed ‘Dongbei longdan.’ The radix et rhizome of G. manshurica was used as traditional Chinese medicine for the treatment of chronic liver diseases for thousands of years (Wang et al. Citation2010). Previous studies on G. manshurica are mainly focused on the resource, pharmacology, and clinical application (Lv et al. Citation2018). However, there have been no genomic studies about G. manshurica. In this study, we reported the complete cp genome and phylogenetic analysis of G. manshurica for the first time, which will contribute to further studies on its genetic research and resource utilization.
The fresh leaves of G. manshurica were collected from the medicinal plant arboretum, Jiangxi University of Traditional Chinese Medicine (Nanchang, China, 28°68′04′′N, 115°75′68′′E), and the voucher specimen was deposited in the herbarium of the Institute of Chinese materia medica, China academy of Chinese medical sciences (HBGP0419). Total DNA was extracted using the DNeasy plant mini kit (QIAGEN). Paired-end reads were sequenced by Illumina HiSeq system (Illumina, San Diego, CA). Approximately 5.21 Gb raw data (34,745,998 reads) were obtained. De novo genome assembly was conducted by NOVOPlasty v3.7 (Dierckxsens et al. Citation2017). The complete cp genome was annotated using GeSeq (Tillich et al. Citation2017) with default sets.
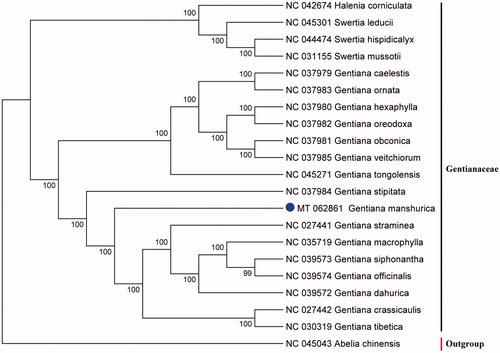
The annotated cp genome was submitted to the GenBank database with the accession number of MT062861. The result showed that the cp genome of G. manshurica was a typical circular structure of 149,185 bp in length, with a large single-copy region (LSC) of 81,347 bp, a small single-copy region (SSC) of 17,268 bp, and a pair of inverted repeats (IRs) regions of 25,285 bp. A total of 130 genes were annotated, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 37.61%. To identify the phylogenetic position of G. manshurica, alignment was performed on the 21 cp genome sequences using MAFFT v7.307 (Katoh and Standley Citation2013), and a maximum likelihood (ML) tree with 1000 bootstrap replicates was constructed by RAxML v8.2.12 (Stamatakis Citation2014; He et al. Citation2020). The ML tree indicated that all the species of the genus Gentiana formed one clade with high support (100%), and the genus Halenia was clustered with genus of Swertia, with 100% bootstrap values (). The cp genome of G. manshurica will provide important information on phylogenetic and evolutionary studies in Gentianaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- He M, Wang X, Zhuang Y, Jin X. 2020. The complete chloroplast genome of Bougainvillea glabra. Mitochondr DNA B. 5(1):889–890.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lv X, Sun JZ, Xu SZ, Cai Q, Liu YQ. 2018. Rapid characterization and identification of chemical constituents in Gentiana radix before and after wine-processed by UHPLC-LTQ-Orbitrap MS(n). Molecules. 23(12):3222.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizer T, Ulbricht-Jones TS, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang AY, Lian LH, Jiang YZ, Wu YL, Nan JX. 2010. Gentiana manshurica Kitagawa prevents acetaminophen-induced acute hepatic injury in mice via inhibiting JNK/ERK MAPK pathway. WJG. 16(3):384–391.