Abstract
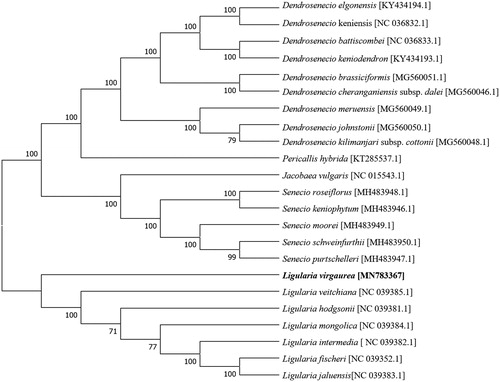
Ligularia virgaurea (Asteraceae) is an indicator species of alpine meadow degradation. Here, the complete chloroplast genome of L. virgaurea was studied. The chloroplast genome of L. virgaurea is 151,185 bp in length. It has a typical quadripartite structure, containing a large single copy region of 83,369 bp, a small single copy region of 18,156 bp, and a pair of inverted repeated regions of 24,830 bp. A total of 132 functional genes were annotated, including eight rRNA genes, 37 tRNA genes, and 87 protein-coding genes. In the neighbour-joining phylogenetic tree based on chloroplast genome, the genus Ligularia clustered together, while L. virgaurea has a slightly more distant relationship with other Ligularia species.
Ligularia virgaurea (Maxim.) Mattf. (Asteraceae: Senecioneae), a poisonous weed, is also an indicator species of alpine meadow degradation on the Qinghai-Tibet Plateau (Wang et al. Citation2008). On the one hand, its population expansion has seriously affected the economic service function of meadow ecosystem. On the other hand, more than thirty species of Ligularia species are used as medicinal materials for the treatment of bronchitis, coughing, pulmonary tuberculosis, and hemoptysis in China (Chen et al. Citation2018). Chloroplast genome of plants is the smallest and most gene-rich genome in each cell, and also the one generally present in the highest copy number (Tonti-Filippini et al. Citation2017). In this study, we sequenced the complete chloroplast genome of L. virgaurea (Genbank accession number: MN783367) based on high-throughput sequencing technology, and the phylogenetic analysis of L. virgaurea and its allies was carried out accordingly.
The samples of L. virgaurea were collected from Laji Mountain, Guide County, Hainan Prefecture, Qinghai Province, China (36.31°N, 101.52°E). The experiment and analysis scheme refers to Wang et al. (Citation2019). Total DNA of L. virgaurea was extracted from the fresh, young leaves (about 2 g) with a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen (Specimen Accession number: XIE2019003) was kept in Herbarium of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (HNWP). Genome sequencing was performed using the Illumina HiSeq Platform 2500 (Illumina, San Diego, CA) at Genepioneer Biotechnologies Inc., Nanjing, China. Approximately 9.41 GB of clean data were yielded. The trimmed reads were mainly assembled by SPAdes (Bankevich et al. Citation2012). The assembled genome was annotated using CpGAVAS (Liu et al. Citation2012).
The complete chloroplast genome of L. virgaurea is 151,185 bp in length. It has a typical quadripartite structure, containing a large single copy region of 83,369 bp, a small single copy region of 18,156 bp, and a pair of inverted repeated regions of 24,830 bp. The two inverted repeated regions are separated by the two single copies. A total of 132 functional genes were annotated, including 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes. The rRNA genes, tRNA genes, and protein-coding genes account for 6.06%, 28.03%, and 65.90% of all annotated genes, respectively. The GC content of the complete chloroplast genome was 37.45%.
Phylogenetic relationships of L. virgaurea, with 22 other species of Tribe Senecioneae (Asteraceae), were resolved by means of Neighbour-joining. Complete chloroplast genomes were aligned using MAFFT (Katoh and Standley Citation2013; online version: https://mafft.cbrc.jp/alignment/server/). The Neighbour-joining tree was built using MEGA 7 (Kumar et al. Citation2016) with bootstrap set to 1,000. In the neighbour-joining phylogenetic tree, the genus Ligularia clustered together, while L. virgaurea has a slightly more distant relationship with other Ligularia species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen X, Zhou J, Cui Y, Wang Y, Duan B, Yao H. 2018. Identification of Ligularia Herbs using the complete chloroplast genome as a Super-Barcode. Front Pharmacol. 9:695
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Tonti-Filippini J, Nevill PG, Dixon K, Small I. 2017. What can we do with 1000 plastid genomes? Plant J. 90(4):808–818.
- Wang J, Cao Q, Wang K, Xing R, Wang L, Zhou D. 2019. Characterization of the complete chloroplast genome of Pterygocalyx volubilis (Gentianaceae). Mitochondrial DNA B. 4(2):2579–2580.
- Wang MT, Zhao ZG, Du GZ, He YL. 2008. Effects of light on the growth and clonal reproduction of Ligularia virgaurea. J Integr Plant Biol. 50(8):1015–1023.