Abstract
Neolamarckia macrophylla (Rubiaceae) is a fast-growing tree with high economic value that is endemic to Sulawesi and Moluccas, Indonesia. Here, we presented the complete chloroplast (cp) genome sequence of N. macrophylla. The complete chloroplast genome is 155,406 bp in size and includes a large single-copy region (86,013 bp), a small single-copy region (18,063 bp), and a pair of IR regions (25,665 bp each). A total of 128 genes were predicted, including 8 rRNA, 36 tRNA, and 84 protein-coding genes. Phylogenetic analysis based on 11 plant species chloroplast genomes indicated that the genus Neolamarckia was monophyletic which clustered as a sister group to Antirhea chinensis. These complete chloroplast genomes can be subsequently used for valuable species research of Rubiaceae.
Neolamarckia macrophylla (Roxb.) Bosser belongs to Rubiaceae and is an important reforestation and timber tree in Southeast Asia. Because of its rapid growth rate and strong adaptability, it has been preferentially selected for planting in forestation initiatives and agroforestry systems. Its wood can be used as the raw material of plywood, board, crate and paper (Qalbi et al. Citation2019), and its bark can be used as a traditional medicine to relieve fever (Pereira et al. Citation2015).
Neolamarckia is a ditypic genus including only two species, namely. N. macrophylla and N. cadamba. Neolamarckia macrophylla is endemic to Eastern Indonesia, from Sulawesi to Maluku (Irawan and Purwanto Citation2014), and now is widely cultivated in many countries, such as Philippines, Malaysia, and India (Pereira et al. Citation2015). At present, the researches on N. macrophylla are mainly focused on wood quality and reproduction (Cahyono et al. Citation2015; Qalbi et al. Citation2019), while its genome and the comparison within Neolamarckia have rarely been investigated.
In this study, we aimed to characterize the complete chloroplast genome of N. macrophylla and explored the phylogenetic relationship with N. cadamba and other species in Rubiaceae family.
Total genomic DNA was extracted from fresh leaves of N. macrophylla collected from Bupon, Luwu, South Sulawesi, Indonesia (S3°14′2.77″, E120°12′6.49″). Additional DNA samples were deposited in the Herbarium of South China Agricultural University (CANT) under the accession number IN-2019-3. A genomic library consisting of an insert size of 300 bp was constructed using TruSeq DNA Sample Prep Kit (Illumina, USA) and sequencing was carried out on an Illumina HiSeq Nova platform (Guangzhou Jierui Biotech). The output was a 6 Gb raw data of 150 bp paired-end reads, further trimmed and assembled using GetOrganelle (Jin et al. Citation2018). The assembled cp genome was annotated using Geseq (Tillich et al. Citation2017) and further manually checked by comparison against the complete cp genome of Neolamarckia cadamba (Genbank accession number: NC_041149).
The complete cp genome of N. macrophylla (Genbank accession number: MN877388) is 155,406 bp in length, consisting of a pair of inverted repeats (IRs: 25,665 bp each), separated by a small single-copy (SSC) region (18,063 bp) and a large single-copy (LSC) region (86,013 bp). The overall GC content of the cp genome was 37.56%. There are 128 genes reported, including 84 protein-coding genes, 8 ribosomal RNA genes, and 36 tRNA genes.
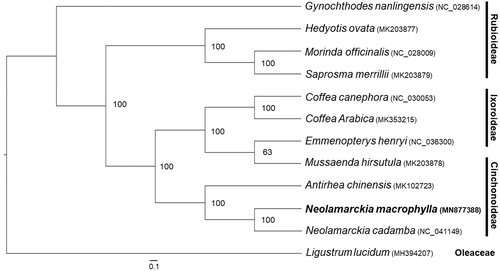
For phylogenetic analysis, a maximum likelihood (ML) tree was constructed based on the complete chloroplast genome sequences of 11 species from Rubiaceae family, with Ligustrum lucidum (Oleaceae) as outgroup. The ML analysis was performed with 1000 bootstrap replicates using RAxML software (Stamatakis Citation2014). The ML tree () showed that N. macrophylla was closest to N. cadamba, the genus Neolamarckia was monophyletic and clustered as the sister group to Antirhea chinensis, and the subfamily Rubioideae was the ancestral group as reported by phylogenetic studies of Rubiaceae with chloroplast sequences (Bremer and Eriksson Citation2009). This finding could serve as a valuable genomic resource for the genetic researches on this important timber and afforestation tree species in future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bremer B, Eriksson T. 2009. Time tree of Rubiaceae: phylogeny and dating the family, subfamilies, and tribes. Int J Plant Sci. 170(6):766–793.
- Cahyono TD, Wahyudi I, Priadi T, Febrianto F, Darmawan W, Bahtiar ET, Ohorella S, Novriyanti E. 2015. The quality of 8 and 10 years old samama wood (Anthocephalus macrophyllus). J Indian Acad Wood Sci. 12(1):22–28.
- Irawan US, Purwanto E. 2014. White jabon (Anthocephalus cadamba) and red jabon (Anthocephalus macrophyllus) for community land rehabilitation: improving local propagation efforts. Agric Sci. 2(3):36–45.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Pereira JT, Hastie AYL, Sugau JB, Chung AYC, Wong N. 2015. The fastgrowing Jabon Merah, Neolamarckia macrophylla (Roxb.) Bosser (Rubiaceae). Annual Report. Sandakan: Sabah Forestry Department; p. 372–375.
- Qalbi N, A’ida N, Restu M, Larekeng SH, Shi S. 2019. Viability test of gamma-irradiated seeds of Jabon Merah (Neolamarckia macrophylla (Wall.) Bosser) from Luwu Provenance: preliminary study. IOP Conf Ser Earth Environ Sci. 343:1:1–5.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.