Abstract
A cyprinid fish Onychostoma macrolepis is endemic to China with high economic values, and its natural resource is decreasing dramatically in recent years. In this study, the complete mitogenome sequence of O. macrolepis from Qinling-Bashan Mountain area was determined using next-generation sequencing. The mitogenome is 16,597 bp in length, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and one non-coding control region with 940 bp in length. The phylogenetic analyses based on whole mitogenome sequences indicated that the genus Onychostoma did not form monophyly, and it was divided into two clades. The present result will provide basic data for the genetic diversity conservation of O. macrolepis.
A cyprinid fish Onychostoma macrolepis (Bleeker) is endemic to China with its large populations distributed in the upstream of Hanjiang River, a branch of the Upper Yangtze River. It inhabits the fast-flowing cold water usually at an altitude of more than 1000 m (Chen Citation2019; Chen et al. Citation2019). Because of hydroelectric project construction in the upstream of Hanjiang River, the wild population of O. macrolepis is decreasing dramatically, and it has been listed as vulnerable (VU) based on the IUCN categories (Jiang et al. Citation2016). To protect its natural resources, the National Aquatic Germplasm Resources Reserve was established in Qinling-Bashan Mountain area (Dong et al. Citation2016). Analysis of the complete mitogenome may contribute to the genetic diversity conservation of this species.
In the present study, the complete mitogenome of one O. macrolepis individual from Qinling-Bashan Mountain area in the upstream of Hanjiang River was sequenced by Illumina HiSeq4000 (Genbank accession number: MT024680). The specimen was collected from Renhe River in Ziyang County of Ankang City (32°23′ N, 108°18′ E), Shaanxi Province, and stored in Qinba Biodiversity Specimen Museum (Voucher Specimen: QBSM20191007). Muscles were fixed in 95% ethanol for DNA extraction, and the whole fish was fixed with 10% formalin. Geneious 11.0.2 was used for annotating the sequenced mitogenome. Protein-coding genes (PCGs) and rRNAs were annotated by comparing with the published mitogenomes of the Onychostoma species (Chai et al. Citation2016).
Totally 17.06 million paired-end reads were obtained for O. macrolepis, and 64,646 reads were used for assembling the complete mitogenome. The newly sequenced mitogenome of O. macrolepis is 16,597 bp in total length with coverage of 578.4 X. Similar to other vertebrate mitogenomes (Shi et al. Citation2018; Chen and Tang Citation2019), the O. macrolepis mitogenome is composed of 13 PCGs (cyt b, ATP6, ATP8, COI-III, ND1-6, and ND4L), 22 tRNA genes, 2 rRNA genes (12S and 16S rRNA), and one control region (D-Loop). Eight tRNAs and ND6 genes are encoded on the L-strand, and the other 29 genes are encoded on the H-strand. 12 out of 13 PCGs start with the regular initiation codon ATG but the COI gene with GTG. Seven PCGs end with the stop codons TAA or TAG, others using the incomplete stop codon (TA– or T–). The D-Loop is 940 bp in length, including terminal associated sequence (TAS), central conserved domain (CD) and conserved sequence block (CSB). The total content of A and T in the mitogenome of O. macrolepis is 55.11%, and 66.70% in the D-Loop. Consequently the mitogenome especially the D-Loop of O. macrolepis shows an AT preference.
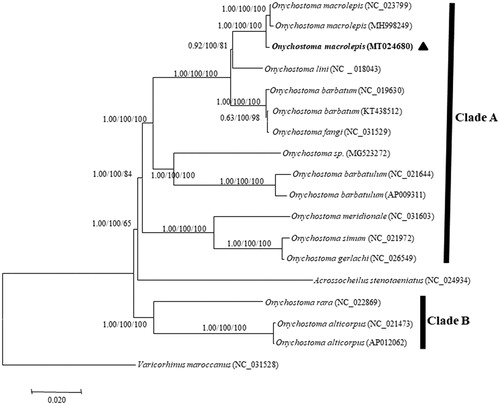
The phylogenetic trees of the genus Onychostoma were constructed based on the newly sequenced and other 17 published whole mitogenomes. Three analysis methods (NJ, ML, and BI) yielded nearly identical topologies (). The genus Onychostoma did not form monophyly, and it was divided into two clades with Acrossocheilus stenotaeniatus being located between Clade A and Clade B, which is congruent with previous studies (Wang et al. Citation2007; Zhang et al. Citation2018).
Figure 1. Phylogenetic tree of the genus Onychostoma reconstructed using neighbor joining (NJ), maximum-likelihood (ML), and Bayesian method (BI) based on whole mitogenome sequences. Values at the nodes correspond to the support values for BI/ML/NJ methods. Solid triangle indicates the newly sequenced mitogenome.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chai A, Zhang J, Cui Q, Yuan C. 2016. Mitochondrial genome of Onychostoma macrolepis (Osteichthyes: Cyprinidae). Mitochondrial DNA Part A. 1:240–241.
- Chen SW. 2019. The study on the biological characteristics of wild Onychostoma macrolepis from Renhe River in Ziyang, Shaanxi province. Shaanxi J Agric Sci. 6:63–66.
- Chen SW, Chen YY, Qu GS. 2019. Analysis and evaluation of nutritional composition of Onychostoma macrolepis in Qinling-Bashan Mountain area. Biotic Res. 2:112–118.
- Chen SW, Tang QY. 2019. The complete mitochondrial genome of big-eye mandarin fish Siniperca kneri (Perciformes: Sinipercidae) from Hanjiang River. Mitochondrial DNA Part B. 1:1298–1299.
- Dong WZ, Wang T, Ma L, Liu Y, Wang LQ, Jiang RM, Ji H. 2016. Artificial reproduction of Onychostoma macrolepis in Qin-Ba Mountains. J Animal Sci Vet Med. 3:23–25.
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL,Xie F, Cai B, Cao L, Zheng GM, et al. 2016. Red list of China’s vertebrates. Biodiversity Science. 24:500–551.
- Shi LX, Zhang C, Wang YP, Tang QY, Danley PD, Liu HZ. 2018. Evolutionary relationship of two balitorids (Cypriniformes, Balitoridae) revealed by comparative mitogenomes. Zool Scr. 47:300–310.
- Wang XZ, Li JB, He SP. 2007. Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol Phylogenet Evol. 42:157–170.
- Zhang C, Cheng Q, Geng H, Lin AH, Wang HY. 2018. Molecular phylogeny of Onychostoma (Cyprinidae) based on mitochondrial genomes. Acta Hydobiologia Sinica. 42:512–516.
