Abstract
Here we sequenced and annotated the complete mitochondrial genome (mitogenome) of Illiberis ulmivora (Lepidoptera: Zygaenidae). This mitogenome was 15,334 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA unit genes (rrnL and rrnS) and a large non-coding region (putative control region). Compared with the putative ancestral arrangement of insects, gene rearrangements were found in two tRNA clusters (IQM and ARNS1EF) of the I. ulmivora mitogenome: changing trnI-trnQ-trnM to trnM-trnI-trnQ, and changing trnS1-trnE-trnF to trnE-trnS1-trnF. The I. ulmivora mitogenome with an A + T content of 81.47% presented a negative AT-skew (−0.029) and a negative GC-skew (−0.206). All tRNAs had a typical secondary cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm. The maximum likelihood analysis based on mitogenomic data highly supported the monophyly of each of Zygaenidae and Limacodidae within Zygaenoidea, and I. ulmivora was sister to the other four Zygaenidae species.
Zygaenidae is the largest family within Zygaenoidea (Insecta: Lepidoptera), with approximate 1,036 species in 170 genera (Van Nieukerken et al. Citation2011). To better understand the evolution and phylogeny of Zygaenidae, we sequenced the complete mitochondrial genome (mitogenome) of Illiberis ulmivora. Adult specimens were collected from Yuzhong County, Gansu Province, China, in June 2018 (104°34´20”E, 36°26´30”N). Samples (voucher no. YZ201806) have been deposited in the State Key Laboratory of Grassland Agro-Ecosystems, College of Pastoral Agricultural Science and Technology, Lanzhou University, Lanzhou, China. The total genomic DNA was extracted from a single specimen using a DNeasy Tissue Kit (Qiagen, Germany).
We obtained the complete mitogenome of I. ulmivora, with 15,334 bp long (GenBank accession number MT075808). This mitogenome contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA unit genes (rrnL and rrnS) and a large non-coding region (putative control region). Compared with the putative ancestral arrangement of insects, gene rearrangements were found in two tRNA clusters (IQM and ARNS1EF) of the I. ulmivora mitogenome: changing trnI-trnQ-trnM to trnM-trnI-trnQ, and changing trnS1-trnE-trnF to trnE-trnS1-trnF. The former tRNA rearrangement is identical to the majority of lepidopteran mitogenomes, whereas the latter is specific to the five Zygaenidae mitogenomes sequenced so for. Gene overlaps were found at three gene junctions and involved a total of 18 bp, ranging in size from 3 to 8 bp. A total of 268 bp intergenic spacers were present in eighteen positions, ranging in size from 1 to 56 bp.
The nucleotide composition of the I. ulmivora mitogenome was biased toward A and T, with an A + T content of 81.47%. This mitogenome presented a negative AT-skew (−0.029) and a negative GC-skew (−0.206) on the J-strand. The rrnL was 1,306 bp long with an A + T content of 84.30%, and the rrnS was 779 bp with an A + T content of 86.01%, as found in most insect mitogenomes. Among the 13 PCGs, the lowest A + T content was 72.44% in cox1, while the highest was 93.08% in atp8. Twelve PCGs started with a typical ATN codon: three (atp8, nad6, nad1) with ATA, three (nad2, nad3 and nad5) with ATT, six (cox2, atp6, cox3, nad4, nad4L and cob) with ATG. The remaining one PCG started with CGA (cox1). Seven PCGs terminated with TAA, and the remaining six stopped with an incomplete stop codon T (cox2). All of the 22 tRNAs, ranging from 62 bp (trnC) to 71 bp (trnK), had a typical cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm.
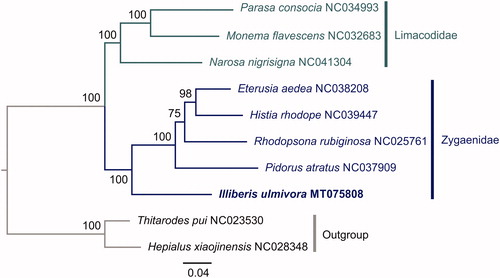
The concatenated nucleotide sequences of 13 PCGs from eight Zygaenoidea species were used in the phylogenetic analysis. Two Hepialoidea species (Hepialus xiaojinensis and Thitarodes pui) were used as outgroups. We conducted a maximum likelihood analysis using RAxML-HPC2 on the CIPRES Science Gateway 3.3 (Miller et al. Citation2010). The monophyly of each of Zygaenidae and Limacodidae within Zygaenoidea was highly supported (). I. ulmivora was placed in the basal, sister to the other four Zygaenidae species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); 2010, New Orleans, LA. p. 1–8.
- VAN Nieukerken EJ, Kaila L, Kitching IJ, Kristensen NP, Lees DC, Minet J, Mitter C, Mutanen M, Regier JC, Simonsen TJ, et al. 2011. Order Lepidoptera Linnaeus, 1758. Zootaxa. 3148(1):212–221.