Abstract
Ficus tikoua Bur. is an important medicinal plant in South China. The complete cp genome sequence is 160,556 bp in length and presented a typical quadripartite structure including one large single copy (LSC) region of 88,620 bp, one small single copy (SSC) region of 20,184 bp, and two inverted repeats (IRs) region of 25,876 bp. The overall GC content was 35.88%. A total of 131 genes were identified, including 86 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. The phylogenetic analysis indicated that F. tikoua was closely related to F. religiosa.
Ficus tikoua Bur. is a woody plant of the Moraceae family, which mainly distributed in south China, India, Vietnam, and Laos (Zhou et al. Citation2018). It’s widely used as a traditional ethnic medicine for treating edema, jaundice, amenorrhea, and bruise, etc. (Cheng et al. Citation2017). Previous studies were focused on its quality standard, chemical composition, and pharmacological activities (Wei et al. Citation2012; Jiang et al. Citation2013). It is well known that the chloroplast (cp) genomes have been widely applied to molecular identification and phylogenetic evolution of plant species (Wang et al. Citation2019). However, up to now, no cp genome of F. tikoua has been reported in NCBI and other databases. Therefore, we characterized and analyzed its cp genome and phylogenetic position, respectively, which would provide new genetic resources for the research of Moraceae.
Fresh leaves of F tikoua were collected from Cangshan Mountains (25°41′4″N, 100°08′31″E) of Dali counties, Yunnan province, China, and the voucher specimen was deposited at the herbarium of Dali University (LJ2020010505). Total DNA was extracted using the DNeasy plant mini kit (QIAGEN). Genome sequencing was performed on an Illumina NovaSeq system (San Diego, CA). Approximately 6.13 Gb raw data (40,916,624 reads) were obtained and assembled by NOVOPlasty (Dierckxsens et al. Citation2017) and then annotated by GeSeq (Michael et al. Citation2017). The annotated cp genome was submitted to the GenBank (accession number MT075555).
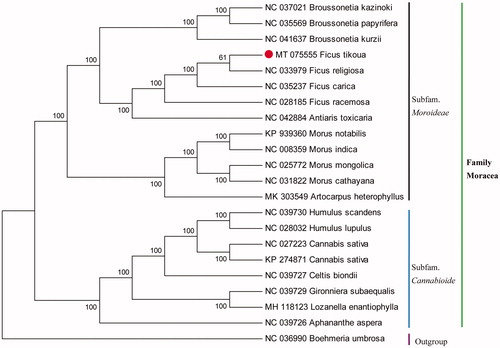
The complete cp genome of F. tikoua is 160,556 bp in length and presented a typical quadripartite structure including one large single copy (LSC) region of 88,620 bp, one small single copy (SSC) region of 20,184 bp, and two inverted repeats (IRs) region of 25,876 bp. The overall GC content was 35.88%. A total of 131 genes were identified, including 86 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. To confirm the phylogenetic position of F. tikoua, 22 cp genomes were downloaded from NCBI and aligned by MAFFT v7.307 (Katoh et al. Citation2002). A neighbor-joining (NJ) tree was constructed using the MEGA 6.0 (Tamura et al. Citation2013), with 1000 bootstrap replicates and Boehmeria umbrosa was used as an outgroup. The results of the phylogenetic analysis indicated that F tikoua was closely related to F. religiosa (). This finding could provide fundamental data for further genetic conservation and phylogenetic studies on Moraceae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cheng Y, Liu SJ, Song JH. 2017. Active components research of Ficus tikoua. Bur.Extracts. Hubei Agr Sci. 1(56):112–114.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jiang ZY, Li SY, Li WJ, Guo JM, Tian K, Hu QF, Huang XZ. 2013. Phenolic glycosides from Ficus tikoua and their cytotoxic activities. Carbohyd Res. 382:19–24.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Michael T, Pascal L, Tommaso P, U E S, Axel F, Ralph B, Stephan G. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol E. 30(12):2725–2729.
- Wang DW, Shi C, Tang HY, He CZ, Duan AA, Gong HD. 2019. The complete chloroplast genome sequence of Docynia indica (Wall.) Decne. Mitochondr DNA B. 4(2):3046–3048.
- Wei S-p, Lu L-n, Ji Z-q, Zhang J-w, Wu W-j. 2012. Chemical constituents from Ficus tikoua. Chem Nat Compd. 48(3):484–485.
- Zhou SY, Wang R, Deng LQ, Zhang XL, Chen M. 2018. A new isoflavanone from Ficus tikoua Bur. Nat Prod Res. 32(21):2516–2522.